Abstract
Background: Cystic fibrosis (CF) is one of the most common indications of preimplantation genetic diagnosis (PGD) for monogenic disorders worldwide.
Aims: The aim of this article was to report a universal and powerful assay easily applicable to all couples requesting PGD for CF irrespective of the CFTR variants involved, in line with recently published CF-PGD guidelines.
Results: A multiplex PCR protocol was developed including the study of the c.1521_1523del mutation with 12 closely linked polymorphic markers. Preliminary workup was performed for 53 couples and the protocol was clinically applied to 31 couples. All couples were informative for 7–12 markers. Of the 31 couples who initiated a PGD stimulation cycle, 17 couples had a baby. Therefore, the take-home baby rate was 60.7% per couple with an embryo transfer (17 out of 28 couples).
Conclusion: This robust, simple, and reliable procedure should allow any couple at risk of transmitting CF to enrol in a PGD programme.
INTRODUCTION
Preimplantation genetic diagnosis (PGD) was first reported in the 1990s as an alternative option to prenatal diagnosis for couples at risk of transmitting a severe monogenic disease or chromosomal disorder to their children.1 The procedure is based on genetic analysis of embryonic cells biopsied from preimplantation embryos obtained through in vitro fertilisation (IVF) techniques. Only embryos free of the disease under investigation are transferred to the mother’s uterus to initiate a pregnancy. PGD in France is strictly regulated by law (only the parental genetic risk can be studied; concomitant aneuploidy screening is not allowed) and is a rare example of an entirely free-of-charge service within the public health organisational framework, thus providing equal access to PGD.2
Cystic fibrosis (CF, OMIM# 219700), a frequent and lethal genetic condition, was the first monogenic disorder to be studied in a PGD clinical case.3 Thereafter, different PGD protocols have been published for CF, ranging from the study of the c.1521_1523del mutation (p.Phe508del) alone4 to more generic strategies based on the study of polymorphic-linked microsatellite markers with or without direct mutation analysis5,6 or karyomapping through analysis of SNP genotypes on microarrays.7 In order to harmonise protocols and procedures for CF-PGD across PGD centres in Europe, specific best practice guidelines have been recently published.8
In line with these guidelines, this paper describes a PGD protocol for CF that presents a notable improvement compared to previously published methods,4-6 as it is now based upon the study of 12 polymorphic markers within or close to the CF transmembrane conductance regulator (CFTR, OMIM# 602421) gene, associated with the direct analysis of the c.1521_1523del mutation.
MATERIALS AND METHODS
Microsatellite Marker Selection
The 9-plex PCR haplotyping approach used previously6 was implemented with the analysis of four additional short tandem repeats (STR) across and flanking the CFTR gene: D7S633 at ~100 kb upstream to the CFTR gene, IVS9TAAA in intron 10, and both CFTRSTR30AC and CFTRSTR15CA microsatellites at ~100 kb and 200 kb downstream to the CFTR gene, respectively.9 All primers were carefully designed and fluorescently labelled to obtain amplicons with different lengths (Table 1) after separation by capillary electrophoresis (Figure 1). Primers for IVS10CA in intron 11 had to be modified to improve electrophoretic peak pattern in this newly designed single-tube PCR protocol (Table 1).

Table 1: Overview of the four additional single tandem repeat markers and the newly designed IVS10CA marker used in the 13-plex preimplantation genetic diagnosis protocol for cystic fibrosis. Their chromosome position, location within the gene or distance to the CFTR gene, type of single tandem repeat and sequence primers, heterozygosity rates, and number of alleles are indicated.
*According to UCSC GRCh38/hg38, December 2013;10 †calculated by typing 129 unrelated individuals. F: forward; R: reverse.
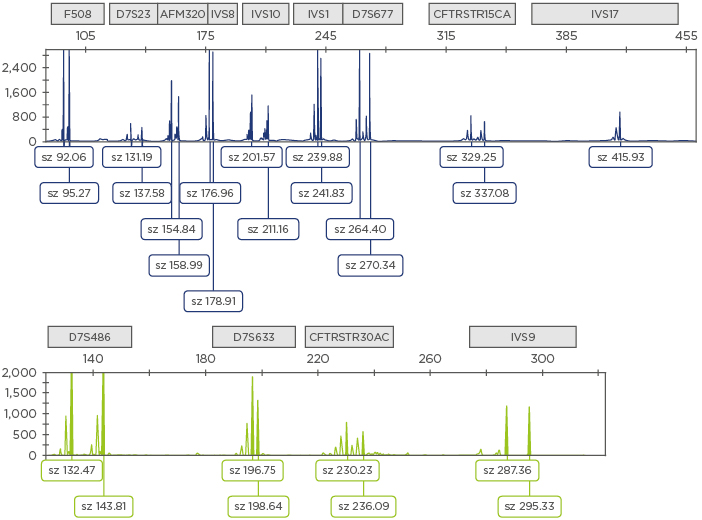
Figure 1: Electropherogram obtained using the generic 13-plex PCR protocol for cystic fibrosis.
Examples of the amplicons are shown for each STR (identified by the squares above the electropherogram). Fam labelled amplicons are shown in blue (top) and Hex labelled amplicons are shown in green (bottom).
STR: short tandem repeats; sz: size.
To estimate heterozygosity rates and to assess informativity for each marker in PGD couples, careful preliminary workup using 13-plex PCR was performed on 100 ng genomic DNA samples from 129 unrelated individuals heterozygous for one severe or large spectrum CFTR gene mutation, including 53 couples enrolled in our PGD programme for CF and 23 individuals referred to our laboratory for CF diagnosis.
Single Cell Multiplex PCR
Before clinical application, to set up the optimal PCR conditions for co-amplification of the 12 STR markers together with the p.Phe508del, >300 single lymphocytes were isolated from different individuals and lysed.6 Reaction mixes were added to the lysed samples to produce a 30 μL final sample. The PCR sample contained 15 µL of 2X Qiagen multiplex PCR master mix and 0.17 µM each of the pairs of primers for p.Phe508del, D7S633, and CFTRSTR30AC; 0.34 µM each of the pairs of primers for D7S486, AFM320vb5, D7S677, D7S23, and IVS10CA; 0.5 µM each of the pairs of primers for IVS1CA and IVS9TAAA; 0.67 µM each of the pairs of primers for IVS8CA and CFTRSTR15CA; and 0.84 µM each of the pairs of primers for IVS17bTA/CA. Thermal cycling consisted of an initial denaturation step at 95°C for 15 minutes, followed by 40 cycles at 94°C for 30 seconds, 58°C for 90 seconds, and 72°C for 60 seconds, then a final extension step at 60°C for 30 minutes. Of the amplified products, 1 μL was run on an ABI 3130XL DNA sequencer and the results were analysed using Genemapper v4.0 software (Applied Biosystems, Foster City, California, USA). For couples carrying variants other than p.Phe508del, informative markers can also be co-amplified with mutation-containing CFTR amplicons to combine mutation detection using a minisequencing approach and linkage analysis.11
The PCR protocols were identical for both single lymphocytes (pre-PGD workup) and biopsied blastomeres from preimplantation embryos (PGD cycles). The IVF part of the PGD procedure has been detailed elsewhere.12
RESULTS
Heterozygosity Rates and Informativity
Heterozygosity rates for the four newly designed microsatellite markers D7S633, IVS9TAAA, CFTRSTR30AC, and CFTRSTR15CA ranged from 76.0–90.7% (Table 1). Among the 53 couples who had a familial pre-PGD workup for CF using the 13-plex PCR protocol, the number of informative markers ranged from 7–12. For the eight couples who displayed only 1–3 fully informative markers using our previously described protocol,6 the study of the four newly designed STR yielded a better informativity with at least one additional fully informative marker.
Single cell amplification and allele drop out (i.e., the random non-amplification or detection of one allele in a heterozygous sample) rates for the different sequences were within the ranges established by the European Society of Human Reproduction and Embryology (ESHRE) PGD consortium best practice guidelines.13 We therefore considered that the updated protocol, detailed in this report, fulfilled the criteria of a reliable clinical PGD method.
Preimplantation Genetic Diagnosis Cycles
From July 2014–December 2017, 31 couples initiated at least one PGD stimulation cycle for CF, including 6 couples with a 50% risk of having a CF-affected child (one CF-affected member, the other being a carrier of a severe or large spectrum CFTR mutation) and 25 couples with a 25% risk (both partners heterozygous for a severe or large spectrum mutation) (Table 2).
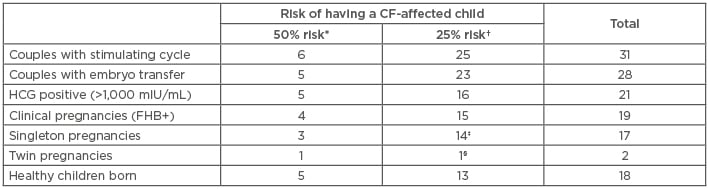
Table 2: Overview of the clinical application of the improved cystic fibrosis preimplantation genetic diagnosis protocol to 31 couples.
*One member of the couple affected with CF, the other heterozygous for a CF-causing or a large spectrum CFTR mutation; †both partners heterozygous for a CF-causing or a large spectrum CFTR mutation; ‡one termination of pregnancy (25 weeks of pregnancy); §spontaneous miscarriage (8 weeks of pregnancy).
CF: cystic fibrosis; FHB+: positive fetal heartbeat; HCG: human chorionic gonadotropin.
Twenty-eight couples (90%) had one or two unaffected embryo(s) transferred and 19 pregnancies (19 couples) with a positive fetal heartbeat were achieved (Table 2). A total of 17 deliveries occurred with 18 healthy babies born. Two adverse outcomes were recorded, including a spontaneous twin pregnancy miscarriage (at 8 weeks) and a termination of pregnancy not related to the PGD indication (at 25 weeks). None of the newborns were diagnosed with CF following testing through the French national newborn screening programme. Therefore, the take-home baby rate is 60.7% per couple with an embryo transfer (17 out of 28 couples).
Discussion
CF is the most common indication for a monogenic disorder at our centre, and one of the most common indications of PGD for single gene disorders reported in the last ESHRE PGD consortium data collection.14 Because >2,000 different genetic variants have been described in the CFTR gene,15 developing and optimising specific single cell PCR tests for each variant is impossible. Novel generic haplotyping technologies, such as karyomapping using SNP arrays,7 are now commercially available for clinical use in PGD and are claimed to be applicable to numerous genotype combinations without prior extensive case-specific workup. However, the cost of the platform is high, the interpretation sophisticated, and the informativity of the biallelic markers may be limited in some cases. PCR-based multiplex assays using informative polyallelic STR markers are still largely used,14 as they represent the simplest strategy allowing the application of a unique genotyping PGD protocol for all couples at risk of transmitting the same monogenic disorder.16
Compared to the 9-plex PCR protocol, which we have previously published, the high number of DNA sequences studied allowed us to obtain conclusive results for all embryos with positive amplification signals, increasing the opportunity to identify unaffected embryos and to achieve a pregnancy. The efficiency and accuracy of our 13-plex PGD protocol is evidenced by the take-home baby rate of 60.7% per couple with embryo transfer. The robust, simple, reliable, and low-cost procedure described in this report should allow the rapid enrolment of any couple at risk of transmitting CF in a PGD programme.








