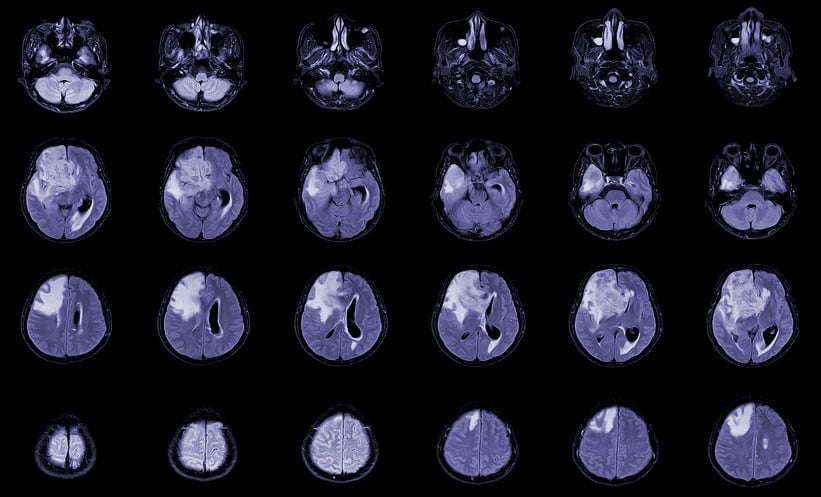
A NEW assessment scheme has been proposed to evaluate the accuracy of automatic tumour segmentation tools for glioblastoma (GBM) using preoperative multi-centre MRI images. The study assessed 74 gross tumour core and T2-FLAIR hyper-intensity segmentations generated by the BraTS-Toolkit and DeepBraTumIA software, alongside 42 manual segmentations performed by neurosurgeons.
The findings reveal that the automatic segmentation tools generally provide accurate results, particularly for tumours with round shapes or clear boundaries. In these cases, the software achieved 100% and 97.06% accuracy for correctly including necrosis and contrast-enhanced tumour areas, compared to 73.68% for manual segmentations. The automatic tools also mistakenly included healthy or non-tumour tissues in only 2.94% and 20.59% of cases, respectively, compared to 10.53% for manual segmentations. T2-FLAIR hyper-intensity segmentations were found to include edema correctly in 88.24% of cases.
However, for tumours with more complex tissue distributions and infiltrative characteristics, manual segmentation outperformed the software tools. In these instances, BraTS-Toolkit and DeepBraTumIA struggled, with the former correctly including necrosis and contrast-enhanced areas in only 50% and 37.50% of cases, respectively. The study also highlighted that MRI image quality significantly impacted automatic segmentation performance for these complex cases.
This research underscores the promise of automatic segmentation tools for simpler tumour types but calls for further refinement to improve performance in more challenging tumour morphologies, especially when image quality is variable.
Helena Bradbury, EMJ
Reference
Cerina V et al. Implication of tumor morphology and MRI characteristics on the accuracy of automated versus human segmentation of GBM areas. Sci Rep. 2025;15:2160.