Abstract
The clinical management of non-small cell lung cancer has shown unprecedented progress into the era of target therapies and immuno-oncology. Despite significant recent achievements in the treatment of these patients, identification of all the clinically actionable alterations required for patient management remains challenging, particularly when dealing with cytological or small bioptic samples. Many investigations have assessed the role of diagnostic tools currently available, including immunohistochemistry and sequencing assays. It is extremely important to be aware of the minimum adequacy criteria for pathology laboratories to ensure correct management of the biological samples in non-small cell lung cancer, including cytological, cell blocks, and histological specimens. In this review, the authors provide a comprehensive overview of the gold standard requirements, processing parameters, and turnaround time for the final integrated report, and additionally outline the values and limitations of the different bioptic strategies.
INTRODUCTION
In accordance with ‘taking care’ of the cancer patient, the management of non-small cell lung cancer (NSCLC) is currently carried out by a multidisciplinary team in which the pathologist plays a pivotal role.1 To allow for patient-tailored testing, appropriate handling of the bioptic material is crucial to allow for the integration of the pathological data with the clinical requests.2-4 Surgery and subsequent radiation or medical therapy is the treatment of choice in NSCLC with loco-regional extension.3 This approach is also possible in selected oligometastatic tumours;5 however, up to 64% of patients with Stage I/II NSCLC, and approximately 76% of those with Stage IIIA NSCLC, relapse and die within 5 years of initial diagnosis.6 The vast majority of patients with advanced NSCLC are considered surgically ‘unresectable’.7-10 In Stage IIIB/IV, the effectiveness of various platinum-based drug combinations are similar, with response rates ranging from 30–40%, a duration of <6 months, and median survival of approximately 12 months.7 In particular, the selection of resistant neoplastic clones by means of somatic evolution and the substantial lack of recognised predictive/prognostic biomarkers are related to treatment failures.11 Lung cancer has become one of the most challenging fields in pathology and oncology over the past decade, contributing to a revolutionary paradigm-shift in patient management.
Starting from the clinical need for prognostic and predictive biomarkers, pathologists have learned to refine their reports. To date, the analysis of epidermal growth factor receptor ([EGFR] on locus 7p11.2) gene mutations, anaplastic lymphoma kinase ([ALK] on locus 2p23.2-p23.1), and ROS proto-oncogene 1 ([ROS1] on locus 6q22.1) rearrangements, and programmed death-ligand 1 (PD-L1) status is capital for clinical decision-making.12-14 In addition to these biomarkers, other actionable gene alterations are under investigation. MET proto-oncogene, receptor tyrosine kinase gene (MET) exon 14 skipping was identified in approximately 5% of NSCLC patients and showed sensitivity to treatment with crizotinib and cabozantinib.13
Activating mutations of EGFR can be observed in 10–16% of lung adenocarcinomas in European patients12-14 and are more frequently observed in young, female, Asian, non-smokers; however, these clinical parameters cannot be used as exclusive selection criteria.15 Several Phase III studies involving patients with NSCLC with sensitising mutations of EGFR showed, compared to standard chemotherapy, a high percentage of approximately 70% of objective responses and a significant increase in progression-free survival of 7–13 months in patients treated with EGFR tyrosine kinase inhibitors (TKI).16-18 The use of EGFR TKI in the front line of the population with mutated EGFR is considered a therapeutic standard and allows for routine research in all new cases affected by NSCLC, particularly non- squamous. The ALK gene rearrangement is present in approximately 3–5% of all pulmonary adenocarcinomas, especially in those with signet ring cell morphology, and predicts the response to ALK inhibitors.19-23 This rearrangement is infrequent in squamous tumours, although it has been reported in mixed adenosquamous forms, and is generally mutually exclusive to EGFR and RAS mutations.24 ALK testing is currently considered mandatory and, for practical reasons, it is usually performed with EGFR testing as a reflex test by immunohistochemistry (IHC) or fluorescence in situ hybridisation (FISH).
The ROS1 gene codes for a tyrosine kinase receptor belonging to the insulin receptor family.25 There are several fusion patterns of ROS1, the most frequent of which is CD74. The translocation of ROS1 is present in 1–2% of the population affected by NSCLC, and in relatively young patients with pulmonary adenocarcinoma or non-smokers in the remaining population.26 There is high homology of the tyrosine kinase domain between ROS1 and ALK, and a preclinical study showed that NSCLC cell lines with translocated ROS1 were sensitive to crizotinib.27 The U.S. Food and Drug Administration (FDA) has approved crizotinib for the treatment of patients with metastatic ROS1-positive NSCLC and is, to date, considered the most appropriate therapy for these patients.28 The latest biomarker that oncologists have claimed as a potential tool for increasing precision medicine is PD-L1 IHC analysis, due to the immunochemotherapy advent.29-34 Other biomarkers under investigation are represented by the analysis of the tumour mutational burden (TMB), mismatch repair, and microsatellite instability status for immunotherapy treatment selection, although they do not represent a global standard of care.35-38 In this scenario, it is extremely important to be aware of the minimum adequacy criteria for molecular pathology laboratories to ensure correct management of the biological samples in NSCLC, including cell blocks (CB), cytological, and histological specimens (Figure 1). Here, the authors illustrate the gold standard requirements, processing parameters, and turnaround time for the final integrated pathology report and outline the values and limitations of the different bioptic strategies.
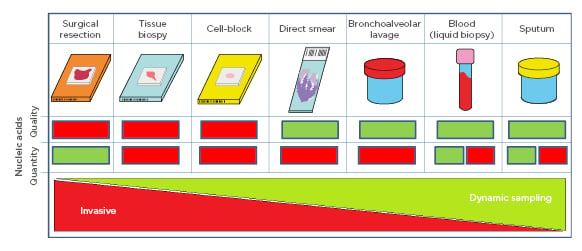
Figure 1: Schematic representation of sample specimens and their quantity and quality of nucleic acid characteristics where green signifies ‘good’ and red signifies ‘bad’. Collection invasiveness decreases dynamic sampling increases from left to right. For blood and sputum, the red box refers to circulating tumour DNA, while the green box refers to genomic DNA.
SAMPLE TYPES
General Considerations
The first task of the pathology laboratory is correct management of the biological samples. This is a crucial step for precise diagnosis and subsequent biological characterisation of the neoplasms. Microhistological analyses are considered a good link between traditional histopathology, first-generation molecular biology, and next-generation sequencing (NGS) techniques. In many European countries, fine needle aspiration (FNA) remains the standard tool for the diagnostic approach to NSCLC both in peripheral pulmonary sites and lymph nodes.39,40 In this setting, the preparation of CB is preferable for molecular testing and requested for the PD-L1 status analysis. For this reason, the rapid on-site evaluation (ROSE) of cytological samples should be performed by a pathologist or a trained cytotechnician, whenever possible. For centrally-located NSCLC, bronchoscopy allows for the direct observation of the lesion to evaluate its characteristics and also to conduct tissue sampling.41 One of the major drawbacks of the histological and molecular characterisation of NSCLC is that the bioptic samples are usually represented by fragments measuring no more than 2 mm in greatest dimension. Furthermore, the analysis of these small fragments is often hindered by necrosis, spread inflammation, and/or crushing artefacts.
For lesions that cannot be assessed using bronchoscopy, a transthoracic core needle biopsy (CNB) may be performed under imaging guidance, for example C-arm cone-beam CT (CBCT). This technique consists of a flat-panel volume CT in which a cone-beam X-ray tube and a flat-panel detector are integrated with a C-arm gantry, enabling both CT and fluoroscopy guidance.42,43 The CBCT virtual navigation systems create a virtual needle pathway to the target nodule and navigates the needle into the target after the operator determines the skin entry site and destination based on pre-procedural CBCT data. The use of 18-gauge semi-automatic biopsy needles with a 17-gauge coaxial needle obtains 1.5 cm-long cores in lesions with a diameter >2 cm. The diagnostic performance may reach 90% sensitivity; however, mean core size is crucial for the success or failure of the procedure.44 Positive predictive value (PPV), negative predictive value (NPV), and overall accuracy in a large series were 100%, 100%, and 87.7%, respectively. Sensitivity was significantly lower for small lesions (57.1%; p<0.01 for lesions ≤10 mm).43 For CNB sampling, the radiologist carries out an average of two passes per nodule, with rare exceptions due to technical issues such as the occurrence of a haemorrhage after the first needle passage.
Transthoracic Core Needle Biopsy Versus Bronchoscopy and Translational Molecular Research
Bioptic fragments from bronchoscopy are immediately formalin fixed and touch preparations to perform ROSE are not reccomended. Fixation should be set in cold (4 °C), 10% neutral formalin for the shortest possible time, typically 6–12 hours for small biopsies. Pathology laboratory technicians should prepare one section for the haematoxylin and eosin (H&E) staining and additional sections for the IHC characterisation of positive cases, using a minimum panel composed of p40, which is more specific than p63, for squamous cell carcinoma, and thyroid transcription factor 1 (TTF1) for adenocarcinoma. Other markers such as synaptophysin, chromogranin, or insulinoma-associated protein 1 (INSM1) should be performed in the presence of neuroendocrine differentiation. In the case of a NSCLC diagnosis favouring adenocarcinoma, subsequent PD-L1, ALK, and ROS1 reflex testing can be completed in 24–48 hours. For CNB, setting up blank slides is encouraged too; the formalin-fixed paraffin-embedded (FFPE) section obtained by CNB can be used to extract DNA for molecular analyses or can be treated to be evaluated by cytogenetic techniques such as FISH. The morphological distinction between neoplastic and benign areas is usually immediate, and necrosis or foci of inflammation can be avoided. The leftover paraffin inclusions should be stored at room temperature. Blank slides should not be used for IHC after a few weeks of storage due to antigenicity loss phenomena; however, they can still be employed for extractive molecular techniques.
The most clinically relevant EGFR mutations concern the deletions of exon 19 or the point mutation L858R in exon 21 (80–90% of all mutations), but even rarer mutations in exon 18 and 21 can predict response to the EGFR tyrosine kinase.45 Different strategies, for either clinical or research purposes, to detect the presence of EGFR gene mutations are employed in molecular predictive laboratories according to the available technology. Sanger direct sequencing, massARRAY® (Agena Bioscience, Inc., Hamburg, Germany), and PCR based techniques still represent the ’gold standard’ methodologies for molecular testing even if they are not the most sensitive.46-49 The College of American Pathologists (CAP), the International Association for the Study of Lung Cancer (IASLC), and the Association for Molecular Pathology (AMP) guidelines recommended that for EGFR gene mutations testing, detection of 50% tumour cells is strongly encouraged, but is rarely achieved in routine practice; when using sensitive detection methods, as little as 10% tumour cell detection is acceptable.50 Currently, for technologies with high sensitivity such as amplification refractory mutation system (ARMS), digital PCR, and NGS applied for gene alterations testing, percentages of tumour cells as low as 1% have been reported.51
ARMS was able to identify a single mutant allele in a background of 100–10,000 wild-type alleles.52 NGS, as demonstrated by Malapelle et al.53 by using a narrow gene panel, was able to detect one mutant allele in a background of 20,000 wild-types.53 A similar sensitivity could be obtained by a digital solid PCR approach.54 In all of these examples, specificity of 100% (no false positive results) were obtained. In order to avoid the risk of false positive and negative results, particularly when considering mutated alleles at low frequencies in paucicellular specimens, careful validation is needed. Secondly, following IHC and instances of ALK Score 2 positive adenocarcinomas, the FISH cytogenetic method evaluates translocation involving EML4 through an intra-chromosomal reversal event. The recommended kit is the Vysis ALK Break Apart FISH Probe Kit (Abbott Laboratories, Abbott Park, Illinois, USA).55 For testing ALK rearrangements the percentages of tumour cells used are not as critical, and areas where tumour cells are not overlapping should be chosen.56 In this subpopulation of patients, however, the appearance of secondary sequence variants in the kinase domain that favour drug resistance have been noted to favour the future advent of alternative diagnostic strategies such as RT-PCR or NGS.57
The test considered to be the gold standard for the determination of translocation of ROS1 is FISH.58,59 IHC methods are currently used as screening tests but, in view of the high false positive rate, tumour samples considered positive in IHC should be tested according to the FISH method.60 Finally, RT-PCR techniques have also been successfully studied.61 Gold standard protocol should produce a 24–48 hour turnaround time for the final diagnostic report of histological type and predictive immunohistochemical markers (i.e., ALK, ROS1, PD-L1). EGFR status should be reported in optimal times of 5–7 days from receipt of the biopsy. For immunochemotherapy, the TMB calculation may be performed using whole exome sequencing (WES) as the standard; however, costs are high, and computational complexity and time make targeted sequencing of pre-customised genes in panels more attractive for routine use. The paraffin block at room temperature is the choice for potential, subsequent NGS profiling. To date, several platforms (SNaPshot Proteomics™ [AVMBioMed™, Limerick, Pennysylvania, USA], MassARRAY MALDI-TOF mass spectrometry, etc.) are available for the massively parallel testing of genetic alterations in NSCLC.62,63 For poor quality specimens, a manual macro- or microdissection enrichment strategy, which can result in highly purified tumour cells for DNA extractions or repeated biopsies, are requisite.64 Enrichment strategies, such as laser capture microdissection or flow cytometric sorting for isolation of tumour cells from small biopsies, should be used cautiously because of their typically low yields of DNA.
Fine-Needle Aspiration and Cell Block
Cytology includes a variety of materials, such as fine-needle aspirations (FNA), bronchial brushing or lavage, sputum, and effusions. The most important parameter is the proportion of tumour cells to non-tumour cells. FNA usually show the highest tumour cell count, in comparison with exfoliative cytology materials.65,66 All materials need critical pre-analytic evaluation to provide tumour cell enrichment for molecular testing.67,68 Samples should be immediately fixed for Papanicolaou staining, using alcohol-based spray or liquid 100% ethanol; alternatively, air-dried preparations may be stained with May-Grünwald-Giemsa (MGG). In the case of ROSE, a Diff-Quik Stain Kit (Polysciences, Inc., Warrington, Pennsylvania, USA) is an option. Especially for brushing and bronchial lavage, an acceptable option is the use of liquid-based cytology.68 Advantages include the elimination of blood in excessively haemorrhagic samples and the possibility to use leftover material for ancillary tests as immunocytochemical staining. Liquid-based preparations may overcome the need for ROSE, reducing artefactual problems related to smear and fixation time. In different studies, EGFR mutation analysis on cytological material gave reliable results. High concordance rates between cytology and histology have been demonstrated in the literature.69 Exfoliative cytological material generally missed a higher proportion of <25% of EGFR mutations.
The use of CB is also suggested because most commercially available kits are validated for FFPE materials.70 The loss of tumour cells and hypocellularity are the two main disadvantages of CB. Molecular tests can be performed on direct smears, liquid-based preparations, and materials stored on filter papers.71 Both MGG and Pap-stained archived slides may be used but air-dried smears are significantly superior to Pap-stained slides in terms of higher EGFR mutation rates. Knoepp and Roh71 suggested the use of Diff-Quik staining because of better preservation of DNA, whereas Killian et al.72 demonstrated DNA degradation in archived Pap-stained smears as a result of alcohol fixation. Contrastingly, there are also studies showing superiority or equivalency of Pap- to MGG-stained slides.73 DNA quality and homogeneity of tumour cells within materials have been shown to be more reliable parameters than quantity.73 CB represent a ‘hybrid preparation’ between cytological and histological specimens, due to FFPE of the FNA material.74 The main advantages of CB include being able to identify the architectural pattern perform ancillary techniques, such as IHC.75,76 Differing from direct smears, CB do not require particular molecular validation as with histological material.77 In order to evaluate the possibility of a significant reduction in neoplastic content and assess the neoplastic cellularity, it is good practice to stain first and use CB sections last.78 This could significantly reduce the risk of false negative results. It is important, particularly in the case of low tumour content, to select neoplastic cells on H&E stained slides to guide tumour cell dissection.74 On the other hand, adopting different serial sections from CB could allow the molecular analysis on the entire cell nucleus.78 An important limitation of direct smears is the low quality of nucleic acids.78-80 To date, a validated cell transfer technique allows the feasibility of molecular testing on DNA extracted from neoplastic cells derived from routine smears.81,82
Another possibility is by liquid-based cytology, in particular, to avoid problems correlated with inadequate triage of aspirated material by untrained clinicians.83,84 In this preparation, the FNA is completely expelled in an alcohol-based fixative, such as CytoLyt® (Hologic, Marlborough, Massachusetts, USA) or CytoRich™ Red (Thermo Fisher Scientific Inc. UK, Leicestershire, UK) solutions, in order to generate a cell monolayer slide.85 Several studies have described the feasibility of liquid-based cytology for molecular purposes.74,85-89 Neoplastic cells are scraped directly into an Eppendorf collecting tube, or obtained by cell lifting with the Pinpoint solution of the Pinpoint Slide DNA Isolation System (Zymo Research, Irvine, California, USA).90,91 A crucial point for molecular analysis is the coverslip of smears in order to scrape tumour cells by a dedicated blade.74 Another possibility to reduce the time of preparation is represented by the ‘freezer method’, in which slides were frozen and a blade was used to remove the coverslip.92 An important issue was the necessity for neoplastic cell enrichment, particularly when low sensitive techniques were employed.77,93
BIOFLUIDS
Sputum and Bronchoalveolar Lavage
Sputum is an important source of nucleic acids, proteins, and other analytes that reflect the status of different organs.94 Different experiences focalised the attention on sputum to investigate EGFR status in NSCLC patients. In a large series (N=50), Wu et al.95 identified a high concordance rate between sputum and tissue samples (74%).95 Hubers et al.96 reported a specificity of 100% but low sensitivity of 50%. In addition to predictive purposes, sputum could also be adopted for diagnostic aims and for secondary prevention.97 Recent evidence suggested the possibility to analyse microRNA (miRNA) in sputum as a non-invasive tool for NSCLC diagnosis.98 In the experience of Bagheri et al.,98 the authors identified expression of miR-223 in sputum as a useful diagnostic biomarker to detect NSCLC patients. In order to increase either sensitivity or specificity, Su et al.99 implemented the analysis of two different biomarkers in sputum (miR-21, miR-31, and miR- 210, and methylation of RASSF1A, PRDM14, and 3OST2 genes). Bronchoalveolar lavage (BAL) is a non-invasive procedure useful for diagnostic purposes in different lung diseases.100 Tuo et al.101 identified the potential role of P16INK4a gene promoter methylation in both BAL and sputum as a diagnostic biomarker for NSCLC, but because of the low sensitivity, it is not suitable as a screening tool. Ren et al.102 showed that SHOX2 and RASSF1A methylation in BAL can increment the detection rate of lung cancer, with high sensitivity and specificity. Kim et al.103 showed the high diagnostic role of five miRNA (miR-21, miR-143, miR-155, miR-210, and miR-372) both in sputum and BAL, notably in the early stage of the disease. However, for these biomarkers, careful attention should be paid to possible artefacts, for example FFPE DNA bisulphite conversion. FFPE DNA bisulphite conversion leads to a challenging methylation sequencing data process due to C→T conversion. Bisulphite sequences are not perfectly complementary to the standard reference genome and, in these cases, special alignment tools are necessary.104
Blood
Due to delays in diagnosis, a high percentage (approximately 70%) of NSCLC patients only have small tissue samples (biopsies and cytological specimens) available for either morphological diagnosis or molecular purposes. 105 To avoid inadequate results, the ‘liquid biopsy’ represents a valid sample to assess the molecular status of EGFR.106 To date, the only analyte approved for EGFR molecular assessment is circulating tumour DNA (ctDNA) extracted from plasma in patients either with diagnosis of advanced NSCLC before any treatment and without the availability of tissue (basal setting), or with resistance to treatment with first- or second-generation TKI for the detection of EGFR exon 20 p.T790M (progression setting).106 The most important issues related to ctDNA regard the low concentration, 0.5% of cell-free DNA, short half-life of approximately 15 minutes, and the modification of concentration during the disease.107,108 For these reasons, the International Association for the Study of Lung Cancer (IASLC) established a statement paper with recommendations for liquid biopsy management.109 In Phase III randomised clinical trials for gefitinib (IPASS) and afatinib (LUX-Lung 3), ctDNA was extracted from serum. The analysis using real-time PCR showed a low sensitivity, 43.1% and 28.6%, respectively.109 An increase in sensitivity, with 100% specificity, was obtained in the Phase IV clinical trial for gefitinib (IFUM) and the Phase III clinical trials for afatinib (LUX-Lung 3 and LUX-Lung 6) in which the analyses were carried out on plasma, 65.7% and 60.5%, respectively.109 Increased sensitivity was obtained by Reckamp et al.,110 using a NGS approach. The authors showed sensitivity of 93.0%, 100%, and 87.0%, and a specificity of 94.0%, 100%, and 96.0% for p.T790M, p.L858R, and exon 19 deletions, respectively, compared with matched tissue samples.110 Malapelle et al.53 showed specificity of 100% and sensitivity of 90.5% by analysing ctDNA extracted from either plasma or serum from each patient (basal and progression settings) by using an ultradeep NGS approach. The same panel was adopted in the experience by Pisapia et al.111 who showed 8% EGFR mutated cases on plasma samples (n=63) in basal NSCLC patients. All the mutations were confirmed by digital PCR.111 In the AURA study, Oxnard et al.112 showed, by using a high sensitivity BEAMing digital PCR (Sysmex Co., Kobe, Japan), different sensitivity and specificity when considering different EGFR mutations, the authors particularly focussed on the lower rate for the EGFR exon 20 p.T790M. Sensitivity was 82.3%, 86.3%, and 70.3%, and specificity was 97.5%, 96.5%, and 69.0% for p.E746_A750delELREA, p.L858R, and p.T790M, respectively.
REQUIREMENTS, PERSPECTIVES, AND FUTURE DEVELOPMENTS
Advanced genome sequence technologies have initiated new perspectives in molecular diagnostics. Novel possibilities, including obtaining multiple genome sequences rapidly and at relatively contained costs, are the result of new approaches to next-generation sequencing, especially compared to conventional sequencing techniques. This makes it possible to search for multiple mutations in the same gene, as well as identify mutations of different genes. Currently, application in routine clinical practice is limited by low distribution in companies and by the high expertise necessary for critical interpretation of genetic data. Therefore, only the availability of series with significant numbers and complex case-mixes can serve as candidates for the proposal of pilot projects that aim to validate NGS platform application in international health systems.113,114 Future projects will investigate NGS methods in pathology, verifying performance indicators such as the efficacy, efficiency, and reproducibility of plasma versus tissue NGS molecular tests. Units of oncology molecular pathology should be able to achieve minimum adequacy criteria for clinical-oncological standards for NSCLC management.
Issues with the biopsy samples (FNA versus transthoracic CNB versus small bronchoscopic biopsy versus CB) may be overcome by interventional contribution of pathologists in radiology or performing a ROSE. Moreover a dedicated technical line to the processing of ‘sensitive’ samples for molecular profiling should be arranged in every oncology molecular pathology unit, with a 24–48 hour turnaround time for a final diagnostic report of histotype and predictive immunohistochemical markers (ALK, ROS1, PD-L1).
The EGFR status should optimally be provided 5–7 days from receipt of the biopsy, keeping leftover material in the archive for NGS profiling. In future developments, the complete integration of traditional pathological techniques with those of molecular biology should result in the formulation of a single report, including all the theranostic factors necessary for the medical oncologists. NGS techniques therefore hold great hope for the future, despite current problems related to standardisation, clinical interpretation of data generated, and reimbursement remaining obstacles to implementation in clinical practice.
The latest biomarker claimed as a potential tool in precision medicine for NSCLC is TMB. This analysis, akin to that of mismatch repair protein expression and microsatellite instability, would require tumour-specific protocols and guidelines for the interpretation.115,116 Larger studies are required to verify whether a high TMB is associated with greater probability of response to immune checkpoint inhibitors and to assess the validity of this new biomarker to represent a real standard of care. Despite the outstanding achievements in the management of lung cancer, and identification of novel biomarkers, the road for precision medicine overlooks a correct histological diagnosis. Definitive therapy for NSCLC should not be undertaken in the absence of tissue diagnosis. Next-generation pathologists are tasked with providing an accurate, specific, and comprehensive diagnosis to enable the clinician to develop an optimal plan of treatment and, to an extent, estimate prognosis. It is capital to manage the tissue samples with parsimony to allow for broader and more accurate analyses. The goal of precision medicine, however, could be achieved only using a holistic approach, where the analyses performed by a pathologist are integrated with all clinical information. This approach would constitute the highway to be travelled for improving survival in NSCLC patients.







