BACKGROUND AND AIMS
Sepsis remains one of the leading causes of death worldwide, with high morbidity and mortality rates.1 Early and appropriate antimicrobial therapy is essential for the clinical outcome2,3 because the survival rate of improperly treated patients decreases by each hour of treatment delay.4 The rapid identification of the causative agent of sepsis is crucial for the patients’ outcome. The Sepsityper® kit (Bruker Daltonik GmbH, Bremen, Germany) is a sample preparation method which enables direct application of matrix-assisted laser desorption ionisation–time of flight mass spectrometry (MALDI–TOF MS) to positive blood culture samples, and obtainment of a microbial identification at species level within 30 mins.5 Steps include the lysis of blood cells, not disruptive for microorganisms, followed by centrifugation and washing steps to obtain a bacterial or fungal pellet. This microbial biomass is suitable for MALDI–TOF MS identification by either ethanol/formic acid extraction or direct smear (rapid Sepsityper), but also for further downstream applications, thus significantly shortening the time to reporting of results.6-8
MATERIALS AND METHODS
In this study, the authors evaluated the rapid Sepsityper procedure with a large collection of routine positive blood cultures (N=5,047) collected from February 2018 to October 2019. Further, the authors investigated the use of the residual bacterial pellet to perform susceptibility testing by the Microscan WalkAway microdilution panels (Beckman Coulter, Brea, California, USA). The Sepsityper method was performed following manufacturer’s instruction.9 Briefly, 200 µL of lysis buffer was added to 1 mL of positive blood culture broth. After vortexing, the sample was centrifuged for 2 mins at 14,000 rpm, and the supernatant was discarded. The pellet was resuspended in 1 mL of washing buffer and centrifuged for 2 mins at 14,000 rpm. The supernatant was removed, and the microbial pellet was used to prepare the MALDI Biotyper target for identification. For a subset of samples (1,648 overall: 1,370 enterobacteria; 98 nonfermenting Gram-negative rods; 83 Staphylococcus aureus; 83 coagulase-negative staphylococci; and 23 enterococci) the residual pellet was then used to prepare the inoculums for the Microscan panels (Neg Multidrug Resistant MIC 1, Pos MIC STA 36, and POS MIC E 37 panels).
RESULTS
Overall, the rapid Sepsityper enabled the direct identification of 4,304/4,848 (88.8%) monomicrobial samples, including 67 genera and 170 species. It showed a very good performance for enterobacteria (98.3%), nonfermenting Gram-negative rods (86.7%), S. aureus (96.2%), and enterococci (93.7%). The missed identifications were restricted mainly to a few groups of microorganisms (streptococci, Corynebacteria, Bacteroides, and yeasts) (Table 1). Among the 200 polymicrobial samples, both species were identified in 62 samples, only one species was identified in 73 samples, and none of the species were identified in 65 samples. Susceptibility testing was successful for 1,549/1,648 samples (94.0%), while for 99 samples the growth in the panel was insufficient, corresponding to the samples that delivered a pellet of poor quality.
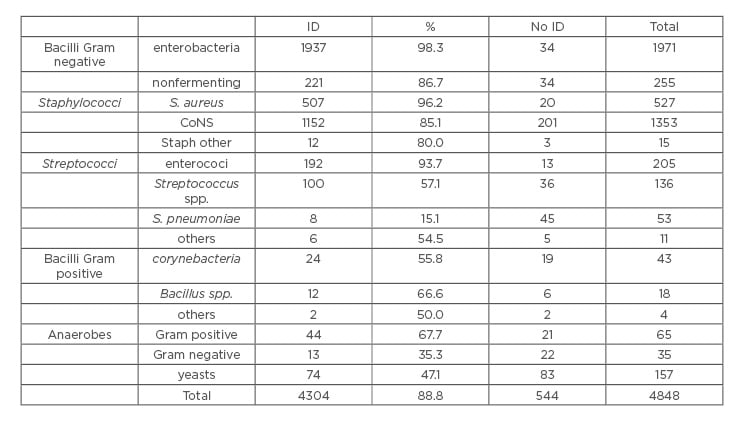
Table 1: Efficacy of Sepsityper® for the identification of different groups of bacteria.
CoNS: coagulase negative staphylococci; ID: identification.
CONCLUSION
In this study, the rapid Sepsityper proved to be a reliable and robust method for bacterial identification directly from positive blood cultures, with an excellent efficacy for the most clinically relevant causative agents of sepsis. It enabled delivery of a result in a very short time, <1 hour for a batch of 10–15 samples, from the harvesting of the positive bottle to the MALDI result. Further, the same bacterial pellet used for the MALDI identification was suitable to set up the antibiotic susceptibility testing, simplifying and speeding up the routine workflow and the time-to-report.








