BACKGROUND AND AIMS
Escherichia coli is the most important bacterial species in Europe, being the main cause of both urinary tract and bloodstream infections.1 The E. coli species consists of at least 10,000 sequence types (ST),2 with ST131 being the dominant cause of quinolone and β-lactam resistant infections throughout Europe.3
MATERIALS AND METHODS
In this study, 968 E. coli isolates from human bacteraemias, faeces, and sewage in 2014 were collected as part of a UK-wide study to assess the reservoirs and origins of E. coli that produce extended spectrum β-lactamase.4 Of these E. coli, 332 belonged to ST131, and a subset of 193 of these isolates (all ST131) were used as representative of ST131 E. coli within the UK for the purpose of this study. Whole genome sequencing of these by Miseq (Illumina, inc., San Diego, California, USA) technology was performed. Core genome (CG) comparison of the ST131 isolates was achieved using blast 2.7.1 on 2,513 CG targets and CG tree generated using Ridom seqsphere (Ridon GmbH, Münster, Germany). The ST131 isolated were scattered throughout the CG phylogenetic tree displaying that the isolates were of diverse genetic makeup within the ST131 sequence type with maximum divergence of <1%.
Each ST131 isolate was tested for susceptibility to bacteriophage. The bacteriophages were isolated from human sewage collected from four countries during April–June 2019: the UK, Kazakhstan, Saudi Arabia, and Bangladesh. Each stock of bacteriophage was collected from similar sized waste-water treatment plants serving similar sized populations in each country. The bacteriophages were simply isolated by first removing the bacteria by centrifugation followed by filtration through a 0.45-micron filter. Bacteriophage were tested for activity by plaque assays on each ST131 isolate and characterised by electron microscopy and sequencing. The E. coli ST131 prevalence was also assessed in each country by random selection of 100 E. coli from the sewage samples and confirmation of the number of ST131 isolates by specific PCR.
RESULTS
The 193 E. coli ST131 isolates represented a broad range of CG multilocus sequence typing (cgMLST) types and were spread throughout the ST131 cgMLST tree. It was found that E. coli ST131 prevalence was dramatically different in the four countries: 11% in the UK, <1% in Saudi Arabia, 4% in Kazakhstan, and undetectable in Bangladesh. However, ST131-specific bacteriophages were highly prevalent in Bangladesh and Saudi Arabia (able to kill 71.75% and 68.20% of UK ST131 isolates, respectively), yet much less common in the UK (31.50%) and rare in Kazakhstan (5.8%)(Figure 1). Electron microscopy revealed that ST131 bacteriophage belonged to several different families including the Siphoviridae, Podoviridae, and Myoviridae.
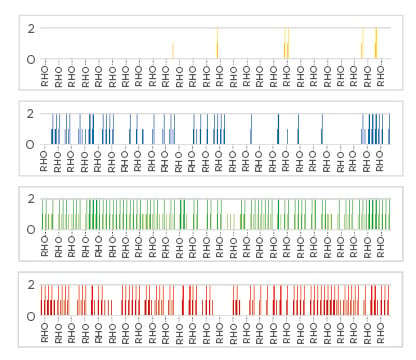
Figure 1: Availability of ST131 bacteriophages in sewage from four different countries.
The number of UK ST131 Escherichia coli strains that can be infected and killed by bacteriophage found in sewage isolated from Kazakhstan (yellow: 5.8%), the UK (blue: 31.5%), Saudi Arabia (green: 68.2%), and Bangladesh (red: 71.8%). Susceptible strains are indicated by vertical coloured lines for each country and assessed for high titre (long lines: >106 plaque forming units/mL) or lower titre (short lines: between 103 and 106 PFU/mL) by plaque assay on individual ST131 E. coli strains.
CONCLUSION
The UK had the highest carriage rates of E. coli ST131, and Bangladesh the lowest.
Thus, ST131 prevalence varies greatly by geographical location. Conversely, sewage from Bangladesh contained bacteriophage that could kill 72% of UK ST131, whereas sewage from the UK could only kill 32% of strains. This strongly suggests that prevalence of ST131 in different countries is controlled by bacteriophage. This difference can be manipulated to target problematic sequence types in one country using a pool of several countries’ bacteriophages. Using a pool of all sites, bacteriophage activity against 91.3% of the 193 ST131 strains can be seen.








