SUMMARY
Altered gut microbiome profiles correlate with anxiety and depression in humans, and work in animal models has identified specific bacterial taxa and/or microbiome-derived metabolites that influence complex emotional behaviours. Intriguingly, many pharmaceuticals, including widely used oral treatments for anxiety and depression, can be chemically modified by microbes in the gastrointestinal tract, which may lead to drug inactivation. The authors highlight the importance of integrating research across microbial culture systems, animal models, and multi-omics analyses of clinical cohorts to gain mechanistic insights into whether microbiome composition determines efficacy, bioavailability, and tolerability of neuropsychiatric medications. This hypothesis, if validated, may have profound implications for personalised drug treatment plans and microbiome-based biomarker development.
THE RECIPROCAL RELATIONSHIP BETWEEN THE GUT MICROBIOME AND MEDICATIONS
The gut microbiome, comprising a staggering 3.8×1013 bacteria along with microscopic fungi, archaea, and viruses in humans,1 plays crucial roles in shaping and maintaining host health. Gut microbes support a wide range of physiological functions including digestion, immune modulation, metabolism, and neuronal signaling. Disruptions in host-microbe interactions are associated with a range of human diseases, such as inflammatory bowel disease (IBD),2 cancer,3 Type 2 diabetes,4 and neurological disorders.5
The gut microbiome is highly dynamic, with community composition influenced by intrinsic factors such as host genetics,6 but also strongly determined by extrinsic/environmental contributors,7 including diet and medication.8 Because diet and drugs are modifiable, understanding the interactions between environmental factors and the gut microbiome offers an exciting and tractable opportunity for development of personalised medicines.
Most pharmaceuticals are administered orally. These substances are either absorbed in the small intestine, where the microbiome is sparse, or pass to the colon, where the densest and most diverse microbial communities reside. Additionally, drugs absorbed in the small intestine may be modified (or not) and secreted back into the intestine, creating new opportunities for exposure to the gut microbiome.9 Consumption of antibiotics, unsurprisingly, has profound effects on the gut microbiome. Acute exposure to a single course of antibiotics can result in the transient reduction or loss of microbial taxa that are important for basic metabolic functions such as carbohydrate fermentation,10 energy production, bile acid transformation,11 and lipid absorption. While most individuals treated with antibiotics experience a rapid recovery of microbiome composition, for some it may take up to 6 months to fully recover their original (pre-drug) microbiome.12 Loss of community stability and, consequently, compromise of normal metabolic functions of the microbiome may lead to opportunistic infections,12,13 deficits in gut barrier integrity,14 weakening of the immune system,15,16 and other unintended consequences. While antibiotics likely have the most profound impact on microbiome function, emerging evidence suggests that other medications may also compromise the microbiome, albeit to a subtler degree.
The vast majority of pharmaceutical drugs were developed against human targets (e.g., proteins, molecules, metabolic pathways), are diverse in structure, and are often consumed for extended periods of time, making it challenging to predict their direct or indirect effects on the microbiome. However, some drug–microbiome interactions have been uncovered. The common Type 2 diabetes medication metformin alters gut microbiome composition in patients, increasing microbial taxa that promote glucose metabolism and thereby increasing its therapeutic effect.17 Methotrexate, a first-line treatment for rheumatoid arthritis, alters microbiome composition in patients and in human microbiome colonised mice, with transplantation of a drug-modified microbiome into drug-naïve mice being sufficient to reduce immune activation.18 The benzisoxazole ring structure in risperidone, an atypical antipsychotic used for schizophrenia and bipolar disorder, is chemically modified by gut microbes, leading to its rapid excretion and thus potentially reducing efficacy and altering dosing regimens in ways that may vary between patients.19
Informed by these findings, there is growing interest in understanding how the gut microbiome may be influenced by, and may influence the efficacy of, various drug classes. Emerging evidence has identified novel microbial transformations of drugs that may alter the intended outcomes of medications.9,20 Given the emerging and likely intricate relationship between gut bacteria and brain function, drug–microbiome interactions in the context of neuropsychiatric disorders represent a particularly interesting area of study. This perspective will first examine how the gut microbiome influences drug metabolism in vivo, drawing from studies in mice and humans with anxiety and major depressive disorder (MDD). The authors will then review known drug–microbiome interactions, primarily through examples beyond neuropsychiatric medications, as these well-characterised cases provide insights into the methodologies needed for future study of microbial metabolism of psychiatric drugs. Finally, the authors will discuss how integrating these approaches can provide an actionable framework for understanding the role of microbial influences on the efficacy and other features of psychiatric drugs.
THE GUT MICROBIOME AND BRAIN HEALTH: INSIGHTS FROM ANXIETY AND DEPRESSION
Anxiety disorders represent the most common class of neuropsychiatric conditions, and are characterised by a persistent avoidance response even in the absence of imminent danger.21 Often co-occurring with depression,22 which is marked by a prolonged loss of interest in activities, anxiety disorders significantly impact quality of life in up to a quarter of the USA and European populations.23,24
In recent years, studies conducted in both mouse models and human cohorts have described a functional role for the gut microbiome in the development of anxiety and depression (Figure 1A). The gut microbiome is stereotypically altered in individuals with anxiety or MDD,25,26 and is speculated to affect symptoms via altered neurotransmitter production,27 inflammation/cytokines,28 the hypothalamic–pituitary–adrenal (HPA) axis,29 vagus nerve,27 and other potential mechanisms. These associations are supported by animal models. Germ-free mice, which are raised without any exposure to microbes, exhibit reduced anxiety-like behaviour, and the reintroduction of a normal microbiome early in life is sufficient to restore anxiety-like traits of standard laboratory mice.30 Transplantation of microbiomes from mice that have experienced chronic stress into naïve recipient mice induces behaviours consistent with depression, and supplementation with Lactobacillus alleviates this effect.31
The gut and brain communicate through various pathways (neuronal, endocrine, immunological) and these interactions involve factors that can be influenced by a diverse array of microbes and their products. For instance, treatment with the bacterium Lactobacillus rhamnosus (JB-1) has been shown to alleviate anxiety and depression-like behaviours in mice.27 This effect occurs through the differential regulation of GABA receptors in the brain and is dependent on the vagus nerve, as vagotomised mice do not exhibit the same behavioural improvements when treated with L. rhamnosus (Figure 1B). Gut bacteria can also produce small molecule metabolites that then travel to the brain and alter cell function: the gut microbial metabolite 4-ethylphenyl sulfate (4-EPS) impairs oligodendrocyte differentiation in mice and increases anxiety-like behaviour.32 In contrast, treating mice with the human commensal Bacteroides fragilis is able to alleviate anxiety-like and autism-associated features (Figure 1C).33
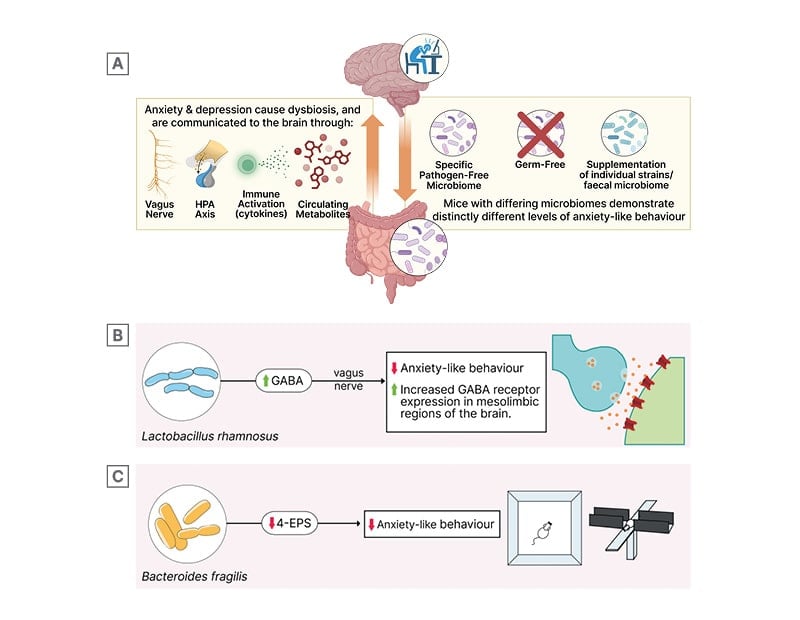
Figure 1: The microbiome and microbially-derived metabolites modulate host nervous system function.
Figure 1A is generated on Biorender.com.
In humans, large cohort studies surveying the microbiomes of depressed patients have revealed stereotypical alterations. Notably, MDD patients often show depletion of genera such as Subdoligranulum and Coprococcus, and an increase in Eggerthella, alongside changes in their metabolomes, particularly increased lipid metabolism.26,34 A recent study integrated microbiome sequencing data from faecal samples of individuals with anxiety and depressive disorders, including those taking medications, to train machine learning algorithms that could successfully predict both the presence of these disorders and medication use based on microbiome profiles alone.35 While effect sizes in human studies remain modest and may necessitate further replication, research to date on the potential pathogenic or protective effects of the gut microbiome in neuropsychiatric conditions represents an exciting frontier of research at the intersection of microbiology, neuroscience, and human health.
Microbiome Modulation of Neuropsychiatric Drugs
Selective serotonin reuptake inhibitors (SSRIs) and selective norepinephrine reuptake inhibitors (SNRIs) are first-line treatments for both anxiety and MDD.36 While these drug classes have been shown to be more effective than placebo for generalised anxiety disorder, their benefits are often accompanied by a therapeutic lag and significant variability in response rates, particularly in terms of long-term acceptability and sustained efficacy.37 Given that SSRIs and SNRIs are orally administered, an important question is whether their interaction with the gut microbiome contributes to the substantial differences in therapeutic acceptability observed across patient populations. Interindividual variations in gut microbiome composition may influence drug metabolism and bioavailability, potentially explaining why some patients respond better to treatment than others.
Recent studies with high-throughput, in vitro culture-based screening systems have revealed extensive drug–microbiome interactions (Figure 2). In one study, researchers exposed 76 individual strains of diverse human gut bacterial taxa to over one hundred commonly prescribed drugs, including medications for anxiety.38 This work found that metabolic reactions were taxon-specific; i.e., Bacteroidetes primarily hydrolysed drugs with ester or amide groups, while most other strains metabolised drugs containing a nitro or azide group. Of note, 10% of the strains chemically transformed anxiolytics, significantly reducing active drug levels in culture, with the SSRI fluoxetine emerging as the most widely metabolised anxiolytic across isolated bacterial strains. Another study incubated different complex communities derived from human faecal samples with drugs used to treat anxiety, again finding that microbiome composition can broadly influence drug metabolism.39 Some communities had the metabolic capacity to degrade specific drugs, while others did not, highlighting interindividual variability in microbiome-driven drug metabolism.
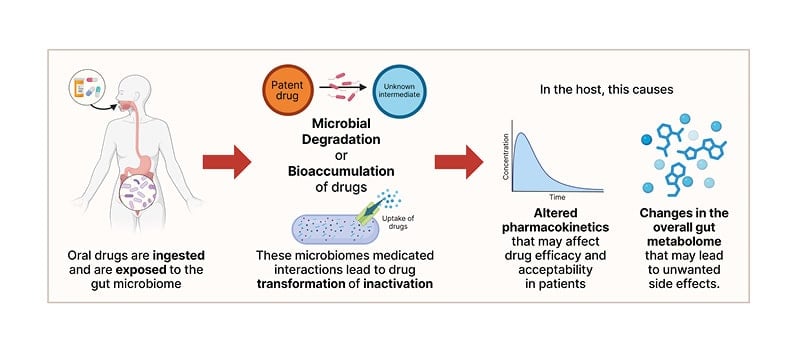
Figure 2: The microbiome modulates drugs, potentially affecting their therapeutic function in the host.
Created on Biorender.com.
Researchers have also leveraged publicly available repositories to develop models to predict drug–microbiome interactions, such as SIMMER (Similarity algorithms that Identify MicrobioMe Enzymatic Reactions)40 and AGORA2 (Assembly of Gut Organisms through Reconstruction and Analysis, version 2).41 SIMMER combines metagenome-assembled genomes, protein homology, and enzyme databases to predict bacterial drug metabolism. This tool identified candidate gut bacterial enzymes, primarily carboxypeptidase G2-like enzymes, with sequence similarity to an environmental enzyme known to hydrolyse methotrexate.40 Experimental testing of strains containing these enzymes confirmed methotrexate degradation. AGORA2 provides a resource for reconstructing metabolic pathways from metagenomic datasets, and incorporates clinical parameters such as BMI and age to facilitate rapid prediction of drug metabolism in epidemiological cohorts.41 Both SIMMER and AGORA2 provide interactive frameworks, allowing researchers to prioritise microbial species, gene products, and pathways of particular relevance for a given disorder and drug class.
While high-throughput screening and large-scale dataset analyses have provided valuable insights, efforts to fully characterise drug–microbiome interactions remain ongoing, with only a few examples to date that have identified products of drug metabolism and even fewer cases tested functionally. For instance, Levodopa (L-DOPA), the first-line treatment for Parkinson’s disease, is degraded by Eggerthella lenta and Enterococcus faecalis.42 These bacterial species were shown to contain enzymes for conversion of L-DOPA into m-tyramine through decarboxylation and dihydroxylation, which may reduce L-DOPA bioavailability and impact treatment efficacy. Another well-known example of a drug–microbiome interaction is 5-aminosalicylic acid (5-ASA), used to treat IBD, whose efficacy is reduced by microbial metabolism.43 By longitudinally monitoring IBD patients on 5-ASA treatment using metagenomics, metatranscriptomics, and metabolomics, researchers identified twelve previously uncharacterised microbial acetyltransferases that were upregulated in non-responders. In vitro assays confirmed that these enzymes acetylate 5-ASA into an inactive form, providing a mechanistic link between microbial metabolism and drug response.
In addition to metabolising drugs, some gut bacteria have been shown to actively transport and bioaccumulate drugs in vitro without modifying their chemical structure (Figure 2).44 Duloxetine, an SNRI, bioaccumulates in diverse gut species, including many from the Firmicutes phylum (Streptococcus salivarius, Clostridium bolteae, Clostridium saccharolyticum, Ruminococcus gnavus, Lactobacillus plantarum, and Lacticaseibacillus paracasei), resulting in altered endogenous metabolism and secretion profiles. Duloxetine modulates Caenorhabditis elegans movement in a dose-dependent manner, and colonisation with the Escherichia coli IAI1 strain that is capable of bioaccumulating duloxetine attenuates this behaviour, highlighting that drug–microbiome interactions can impact behavioural outcomes.44
Finally, the gut microbiome can regulate host drug transporters, thus influencing pharmacokinetics. Differences in microbiome composition, such as between conventionally-raised and germ-free animals, alter the expression of the efflux transporter P-glycoprotein (P-gp/ABCB1),45 which may contribute to pharmacokinetic variability for P-gp substrate drugs, including the SSRI sertraline and the antipsychotic risperidone. However, whether degradation, modification, bioaccumulation and/or altered transport of SSRIs or SNRIs impact anxiety or depression-like behaviours in mammalian model systems remains unexplored to date, defining a frontier of future research.
TOWARDS A HOLISTIC UNDERSTANDING OF DRUG–MICROBIOME INTERACTIONS
While microbial cell culture-based experiments offer rigorous insights into drug–microbiome interactions, these systems are unable to capture the physiology of an organism and its associated microbiome, with studies in freely behaving animals required to advance this research toward understanding effects on emotional behaviours. Recent in vitro findings have also revealed that reductions in drug levels do not necessarily indicate microbial metabolism.20 Abiotic factors, including spontaneous degradation, ion suppression, surface adsorption, and bioaccumulation, can have strong effects on drug activity.
To ensure reproducible and clinically relevant results, it is important to test drug–microbiome interactions within their native host context, minimising artefacts introduced by culture conditions. Moreover, it is possible that long-term medication use can reshape the gut environment and microbiome composition, which then secondarily influences symptoms or treatment outcomes, though this concept remains hypothetical in the absence of empiric evidence. Disentangling these factors requires an integrated approach, combining multi-omics analyses of diverse human cohorts with rodent models or non-human primate models that are amenable to experimental approaches to define functional outcomes. Given that microbial bio-transformations largely fall within a defined set of reaction types, such as reduction, hydrolysis, decarboxylation, and dealkylation, identifying overarching principles governing these transformations may be feasible. Leveraging large-scale machine learning models trained on high-resolution microbiome and metabolomics datasets could offer a powerful strategy to predict drug modifications and their downstream effects, ultimately guiding the design of more precise and effective therapeutic interventions.
As our understanding of drug–microbiome interactions becomes more refined, the development of predictive frameworks for drug efficacy and tolerability based on an individual’s symptoms, lifestyle, medication history, and microbiome status will be increasingly feasible. Such tools could one day help tailor pharmacological treatments to maximise therapeutic benefit, ultimately advancing precision medicine. It is conceivable that gut microbiome variations explain inter-individual responses to numerous classes of oral drugs, beyond those for neuropsychiatric conditions, and potentially even injectables via microbiome modulation of immune profiles (e.g., immune checkpoint inhibitors)46-48 and metabolic states (e.g., weight loss drugs).49,50 Identifying microbiome-based markers that quantitatively predict variance in drug response in defined patient populations may streamline drug discovery and development, improve efficacy rates and response times, and reduce side effects.