Abstract
The field of reproductive health is progressing rapidly from traditional non-molecular technologies based on visual microscope-based techniques to the latest molecular technologies, that are more accurate, objective, and efficient, and some of which are less invasive. Genome-wide technologies have been applied at different stages of the reproductive health lifecycle, such as preimplantation genetic testing, prenatal and postnatal testing, and preconception carrier screening. Next-generation sequencing is currently the platform of choice when it comes to preimplantation genetic testing, and analysis using cell-free DNA offers a potential non-invasive alternative to current methods. Molecular tests of endometrial receptivity identify the optimum timing for embryo implantation, thereby improving in vitro fertilisation (IVF) success rates for patients with recurrent implantation failure of endometrial origin. In the prenatal and postnatal settings, new technologies, such as microarrays and next-generation sequencing, have increased the diagnostic yield and fuelled the rate of discovery of new genetic syndromes. Expanded carrier screening panels have replaced multiple single-gene tests with a single assay and have been shown to be more effective at identifying carriers of genetic disorders. These innovations are accompanied by new challenges regarding their implementation and use. Patient access to new technologies varies greatly and several factors have been identified as barriers to uptake. Genetic counselling has become increasingly important as the amount of genetic information provided by these technologies continues to rise. This review discusses specific challenges associated with traditional non-molecular and older-generation molecular techniques in reproductive health, and suggests potential solutions provided by recent advances in genetic technologies.
INTRODUCTION
In recent years, the introduction of multiple new technologies has dramatically changed the field of reproductive health (RH), with significant implications for clinical practice at all stages of the RH lifecycle. Older technologies are generally based on visual, microscope-based techniques. These techniques are mainly used for diagnostic purposes and are based on invasive sampling; however, they are often limited by poor turnaround time, reproducibility, resolution, and accuracy, as well as high costs. In recent years, there has been a shift away from conventional microscope-based methods towards molecular techniques that are often faster, more reproducible, more efficient, and have increased resolution. These innovations have advanced our understanding of infertility and genetic diseases and have the potential to reduce the diagnostic odyssey and inform patient decision-making in light of the increased genetic knowledge available. In this review, the authors explore specific challenges associated with traditional non-molecular techniques at different stages of the RH lifecycle and describe the potential solutions provided by the latest molecular technologies. A top-line overview of the review is presented in Table 1.
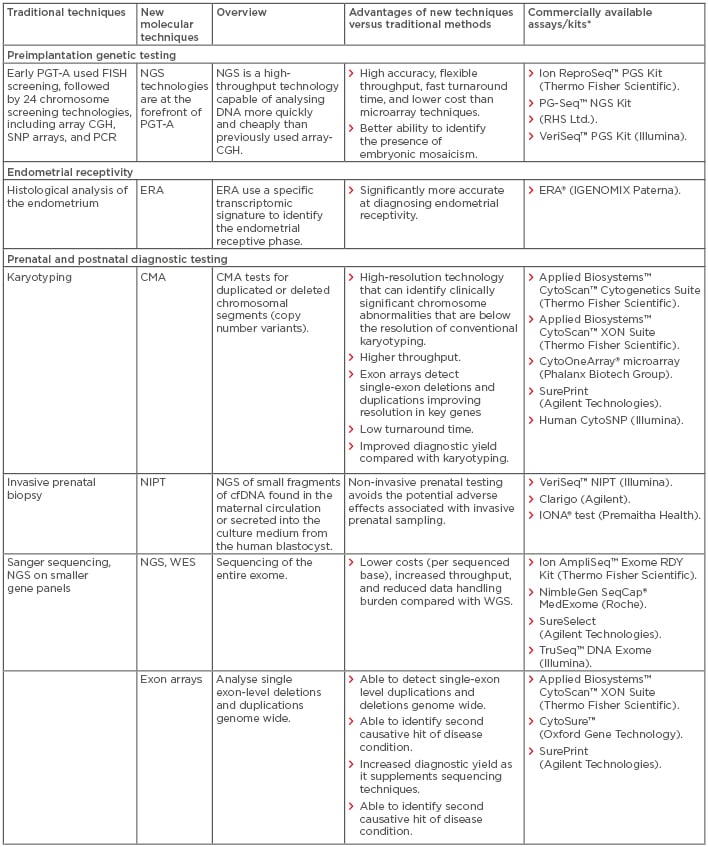
Table 1: Comparison of new molecular techniques in reproductive health with traditional techniques: Overview, advantages, and commercial availability.
*All manufacturers’ websites were last accessed in September 2018.
cfDNA: cell-free DNA; CGH: comparative genomic hybridisation; CMA: chromosomal microarray; ECS: expanded carrier screening; ERA: endometrial receptivity array; FISH: fluorescence in situ hybridisation; NGS: next-generation sequencing; NIPT: non-invasive prenatal testing; PGT-A: preimplantation genetic testing for aneuploidy; SNP: single nucleotide polymorphism; WES: whole-exome sequencing; WGS: whole-genome sequencing.
PREIMPLANTATION GENETIC TESTING
Current Preimplantation Genetic Testing in Trophectoderm Biopsies by NGS Allows More Accurate Detection of Mosaicism. This is an Invasive Approach, but NGS Could be also Applied to Non-invasive Aneuploidy Testing of the Embryonic Cell Free DNA Released to the Media
As women age, their fertility declines and there is an increased risk of numerical and structural chromosomal abnormalities in their oocytes, which can lead to implantation failure, early pregnancy loss, congenital birth defects, or severe chromosomal diseases, such as Down’s and Patau syndromes.1,2 Aneuploidy, the presence of an abnormal number of chromosomes that is not an exact multiple of the usual haploid number, is the most common genetic abnormality and accounts for approximately 50% of miscarriages. More than half of the embryos produced by in vitro fertilisation (IVF) are aneuploid.1
The process of detecting numerical or structural chromosomal abnormalities for the purpose of embryo selection is generally referred to as preimplantation genetic testing for aneuploidy (PGT-A), previously known as preimplantation genetic testing. PGT-A was introduced in the 2000s to increase implantation and pregnancy rates, as well as decrease miscarriage rates, the risk of aneuploid offspring, and the time to conceive.3,4 Early PGT-A used fluorescence in situ hybridisation (FISH) screening. However, data from several studies have been used to question the efficacy of FISH screening,5-8 which is restricted to analysing a limited number of chromosomes.9 In recent years, PGT-A using FISH screening has been replaced by next-generation sequencing (NGS)-based and chromosomal microarray techniques.10 PGT-A assesses the whole chromosome complement (24 chromosomes) and can be carried out with various genetic platforms, such as chromosomal microarrays,11 which detect all mitotic and meiotic abnormalities present in one cell or a group of cells. Chromosomal microarrays can be used not only for testing chromosomal aneuploidy but also to detect unbalanced translocations and other structural chromosomal abnormalities.12
A limitation of PGT-A is the presence of chromosomal mosaicism within the blastocyst, whereby the cells analysed may not be representative of the chromosomal status of the entire embryo, potentially resulting in misdiagnosis.13 Mosaic embryos can develop into healthy euploid newborns, but are associated with significantly poorer implantation and ongoing pregnancy rates and more frequent miscarriage compared with euploid embryos.14-18 Mosaic embryos are not commonly recommended for transfer, but they may be considered as an exception in the absence of euploid embryos and when there is no option for undergoing further cycles of IVF with PGT-A. This approach requires caution, special consent from the patient, and obligatory follow-up with prenatal testing.19-21
Current PGT-A techniques require an invasive embryo biopsy, which has been proposed to reduce embryo quality after cleavage-stage biopsy but not after trophectoderm biopsy.22 The long-term safety of embryo biopsy is unclear in humans, but animal studies have indicated a possible detrimental effect on neural and adrenal development.23-25 The biopsy procedure is technically challenging and requires an experienced embryologist, which, together with the cost of testing multiplied by the number of embryos produced by the couple, significantly increases the overall costs of clinical PGT-A cycles.26,27 An easy-to-use, affordable, non-invasive PGT-A tool may avoid the need for embryo biopsy and improve patient access to genetic testing.
Molecular and Non-Invasive Techniques Have the Potential to Change the Approach to Preimplantation Genetic Testing
Whole-genome PGT-A technologies have been developed, including chromosomal microarrays, multiplex quantitative PCR, and NGS.26 NGS technologies are at the forefront of PGT-A, with the advantages of higher resolution, flexible throughput, and lower cost.16 Furthermore, NGS allows for large numbers of samples to be analysed at the same time; as a result, the per-embryo cost of testing is reduced by more than one-third compared with widely used microarray-based techniques and should, therefore, improve patient access to preimplantation testing.27 NGS technologies may also allow for simultaneous evaluation of single-gene disorders and chromosomal aberrations with comprehensive aneuploidy testing from the same biopsy sample without the need for multiple platforms.28
Compared with other PGT-A methods, NGS also provides a more accurate copy number for each chromosome and is, therefore, better able to identify the presence of mosaic aneuploidy within the blastocyst, since undetected aneuploidy may increase the risk of first trimester pregnancy loss.16 The improved detection of mosaic aneuploidy afforded by NGS could help to identify embryos at high risk of miscarriage.16
To avoid the potential damage caused by embryo biopsy, procedures are being developed that analyse embryonic cell free DNA (cfDNA) released by the blastocyst to the culture media as a non-invasive approach, although these are still considered to be experimental. The reliability of this approach is dependent on whether the chromosomal abnormalities in the cfDNA reflect the anomalies found in the embryo. The concordance between cfDNA and biopsy DNA is highly variable, potentially as a result of different methodological approaches and maternal DNA contamination.9,13,29-31 Optimisation of a PGT-A method using cfDNA could mitigate the potential adverse effects associated with embryo biopsy.29 These approaches are less technically demanding and more cost-effective compared with embryo biopsy so could, therefore, increase patient access to PGT-A.13 There are a number of commercially available kits, including the Ion ReproSeq™ PGS Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA), PG-Seq™ NGS Kit (RHS Ltd., Adelaide, Australia), and VeriSeq™ PGS Kit (Illumina, San Diego, California, USA).
Preimplantation genetic testing for monogenic (i.e., single gene) disorders (PGT-M) is aimed at detecting Mendelian genetic diseases in the embryo and has been applied in >200 disorders to date.32,33 PGT-M uses a variety of methods; the traditional approach is single-cell PCR with linkage analysis of the short tandem repeats related to the pathogenic locus, which requires an individualised design for each disease. However, this process takes 3–4 months and is very costly. More recently, genome-wide, high-throughput methods, such as single nucleotide polymorphism microarray and NGS, have been used.34,35,36
ENDOMETRIAL RECEPTIVITY FOR IMPLANTATION
Histology Does Not Accurately Predict Endometrial Receptivity
Endometrial receptivity issues can result in embryo implantation failure.37 After approximately 5 days of progesterone exposure, the endometrium acquires a receptive phenotype known as the ‘window of implantation (WOI)’, which lasts for approximately 1–2 days and permits implantation of the blastocyst. For >60 years, histological evaluation has been widely used to evaluate endometrial receptivity. Nevertheless, evidence from two randomised studies has indicated that histological evaluation lacks the precision and accuracy to be used as a predictor of endometrial receptivity.38,39
Current histological methods for assessing endometrial receptivity use a ‘one size fits all’ approach based on the assumption that the WOI is consistent in all women.40 However, there is evidence to suggest that some patients diagnosed with recurrent implantation failure (RIF) of endometrial origin should not be categorised as having a pathologic condition but as patients who would benefit from personalised embryo transfer (pET) timing because their endometrial timing is different.41 Other factors that may contribute to RIF are an altered pattern of the microbial endometrial ecosystem or of uterine contractions during embryo transfer.42 Providing a personalised evaluation of endometrial receptivity could improve reproductive outcomes, particularly in patients with RIF.
Endometrial Receptivity Array Can Improve In Vitro Fertilisation Success Rates
An endometrial receptivity analysis (ERA) is a molecular diagnostic tool that uses the specific transcriptomic signature to identify the endometrial receptive phase. The approach is more accurate than traditional histological methods and overcomes issues of intraobserver variability that result from difficulties distinguishing pre-receptive, receptive, and post-receptive histological phenotypes.43
A recent study provided the first evidence of the efficacy of ERA as a diagnostic tool for diagnosing endometrial receptivity in the context of euploid embryo transfer. In RIF patients with a displaced WOI diagnosed by ERA, implantation rates (73.7% versus 54.2%, respectively) and ongoing pregnancy rates (63.2% versus 41.7%, respectively) were higher in patients who received pET versus patients without pET.44 A commercial ERA is available from IGENOMIX Paterna (Valencia, Spain) (Table 1).
A new tool is under development that can predict endometrial receptivity status based on a gene expression analysis, using combined quantitative reverse transcription PCR. The ER Map®/ER Grade® test was validated in endometrial biopsy samples obtained at luteinising hormone (LH) surge+2 days and LH surge+7 days in fertile women in a natural cycle and at WOI in patients in a hormone-replacement therapy cycle. Expression analyses revealed significantly different levels of expression of 85 of 184 genes involved in endometrial proliferation and the maternal immune response associated with embryonic implantation when comparing the LH surge+2 days and LH surge+7 days samples (p<0.05).45 The authors noted that further studies need to be conducted to evaluate the efficacy of this tool, including non-selection studies and randomised controlled trials.
PRENATAL AND POSTNATAL DIAGNOSTIC TESTING
Karyotyping Fails to Identify Sub-Microscopic Chromosomal Anomalies in Prenatal and Postnatal Diagnosis
Prior to the development of molecular techniques, detection of chromosomal abnormalities for prenatal and postnatal diagnosis largely involved conventional karyotyping of cultured live cells.46 This method has practical limitations, including the reliability of successful cell culture with failure rates of 10–40%, high turnaround times, and the technically demanding nature of the procedure, which requires specialist training.47
The resolution of chromosomal analysis using karyotyping is limited by what can be seen under a microscope, which typically is 5–10 Mb in size.46 The majority of chromosomal anomalies identified in early pregnancy are aneuploidies, which are detectable with conventional karyotyping.48 However, rare genetic syndromes are often caused by submicroscopic imbalances.49 Clinically significant copy number variations (CNV) not identifiable by standard karyotyping are estimated to occur in 1.0–1.7% of routine pregnancies.50 CNV are segments of DNA that are present in a different number of copies in an individual compared with a reference genome.46 Studies have supported the causative role of CNV in some individuals in a variety of conditions, including malformations or dysmorphisms from congenital anomalies, intellectual disability and developmental delay, epilepsy, cerebral palsy, and neuropsychiatric disorders.46
New Genetic Technologies Have Improved the Diagnostic Yield in Prenatal and Postnatal Genetic Testing
There are multiple technologies for genetic testing, each with their own purpose and all fulfilling different and important roles in developing a greater understanding of reproductive outcomes. New genetic techniques, such as chromosomal microarray (CMA), have replaced karyotyping as the preferred genomic diagnostic testing methodology for the identification of chromosomal abnormalities because they are faster and provide increased resolution. Furthermore, these techniques are able to detect abnormalities that would be lost during cell culture since they are performed on DNA extracted from chorion villus material, amniotic fluid, or blood samples.
CMA has several advantages over karyotyping in both prenatal and postnatal testing and are becoming the preferred diagnostic tests in these settings.49 CMA is a high-resolution technology that can identify clinically significant chromosome abnormalities that are below the resolution of conventional karyotyping.48 The primary aim of CMA is to identify CNV,46 with pathogenic CNV detected in 5.6% of fetuses with isolated anomalies and 9.1% of fetuses with multiple anomalies.49
Several studies have demonstrated the value of CMA in prenatal and postnatal testing in clinical practice.49,51,52 Increased nuchal translucency is associated with common fetal aneuploidies and fetal genetic disorders, syndromes, and structural defects. In a study examining genetic imbalances in 94 fetuses with increased nuchal translucency, CMA detected pathogenic CNV in 12.8% of cases. Eleven (73.3%) of the cases with imbalances detected by CMA were ≤10 Mb (range: 0.4–5.5 Mb) and were unlikely to have been detected by karyotype analysis.49 Limiting CMA testing to pregnancies with a risk above 1 in 100 or 1 in 50, as proposed in local testing models in Denmark, would have led to a significant number of pathogenic CNV being missed.52 In one medical centre, the introduction of CMA in place of karyotyping led to an additional yield of submicroscopic pathogenic chromosomal aberrations; specifically, there was an increase of 3.6% in fetuses with ultrasound anomalies and 1.9% in fetuses without ultrasound anomalies.51
CMA has some limitations, including the inability to detect balanced translocations and, for CMA without single nucleotide polymorphisms, polyploidy, which could be of clinical significance.46,48 However, the increased detection of clinically significant CNV too small to visualise using conventional karyotyping, as well as greater throughput, outweighs its loss of detection of balanced karyotypic abnormalities.46 CMA is more expensive than karyotyping but cost–benefit analyses have indicated that CMA is cost-effective when used for prenatal diagnosis of an anomalous fetus.53 Testing with CMA is a guideline-recommended option in both the prenatal and postnatal settings.48,54 In many countries, CMA testing is the primary analytical technique used in the testing of all prenatal and postnatal samples.
More recently, the emergence of NGS technologies has resulted in the development of non-invasive prenatal testing (NIPT) assays aimed at detecting fetal aneuploidies by the analysis of circulating free DNA (cfDNA) in maternal plasma.55 NIPT tests differ in their methodology and several different assays are available. NIPT is a well-established option for screening for trisomy 21, 18, and 13, as well as other selected chromosomal abnormalities.56 NIPT can be conducted as early as 10 weeks gestation, is highly accurate, and procedurally safe for both the mother and fetus.57 Commercially available CMA include Applied Biosystems™, CytoScan™, Cytogenetics Suite (Thermo Fisher Scientific); CytoOneArray® microarray (Phalanx Biotech Group, Hsinchu, Taiwan); SurePrint (Agilent Technologies, Santa Clara, California, USA); and Human CytoSNP (Illumina®) (Table 1). Commercially available NIPT include VeriSeq NIPT (Illumina®), Clarigo (Agilent Technologies), and IONA test (Premaitha Health, Manchester, UK) (Table 1).
Whole-Exome Sequencing Has Driven a Rapid Increase in the Detection of Genetic Diseases
The most recent development in prenatal and postnatal genetic diagnostic testing is whole-exome sequencing (WES), which uses NGS technology. DNA containing protein-coding exons comprises 1–2% of the genome but >85% of all disease-causing mutations.58 The development of WES has resulted in a rapid increase in the detection of genetic diseases. WES of 250 patients with disease phenotypes suggesting genetic causes identified the underlying genetic defect in 25% of patients,59 which is higher than the diagnostic yields of other genetic tests, such as karyotyping (5–15%)60 and CMA (15–20%).46 WES also has lower costs, increased throughput, and reduced data handling burden compared with whole-genome sequencing (WGS) and has, therefore, been the preferred technique used in genetic diagnostic testing. However, unlike WGS, WES is not currently able to detect all types of genetic variation, including small copy number changes, low-level mosaicism, structural chromosome rearrangements or trinucleotide repeat expansions, and variants in non-coding regions.58 Commercially available WES currently include Ion AmpliSeq™ Exome RDY Kit (Thermo Fisher Scientific), NimbleGen MedExome (Roche, Basel, Switzerland), TruSeq DNA Exome (Illumina), and SureSelect (Agilent Technologies) (Table 1).
Exon-Targeted Arrays Offer a High-Sensitivity Approach to Genetic Diagnostic Testing
CMA is used to test for genetic disorders that result from submicroscopic deletions or duplications that affect multiple genes but is not best suited for the detection of deletions or duplications with a genomic loci smaller than a single gene.61 Exon-targeted arrays can detect intragenic CNV <1,000 bp, including those that affect only a single exon. Exon arrays can focus on known or candidate disease genes, or target the entire genome with increased focus on clinically relevant genes. This high-sensitivity approach is used to supplement WES and has driven the discovery of novel disease genes. As a result, the molecular diagnosis for conditions with known disease-associated genes has increased, enabling better genotype–phenotype correlations and improved variant allele detection.62 This technology is an efficient, sensitive, and cost-effective method for genetic diagnostic testing.63 Commercially available exon arrays currently include Applied Biosystems™ and CytoScan™ XON Suite (Thermo Fisher Scientific), CytoSureTM (Oxford Gene Technology, Begbroke, UK), and SurePrint (Agilent Technologies) (Table 1).
Whole Genome Sequencing Has the Potential to Capture All Classes of Genetic Variation
In the past, genetic diagnostic tests have focussed on targeted sequencing of candidate genes, requiring prior knowledge of clinical phenotypes caused by mutations in specific genes. This was problematic because many phenotypes or genetic disorders may have highly variable phenotypes or are caused by mutations in multiple genes. The development of NGS technologies that enabled sequencing of the entire genome has resulted in unprecedented increases in speed and reductions in the cost of genetic investigations.64 WGS provides a base-by-base view of all genomic alterations, including single nucleotide variants, copy number changes, and structural variations, and, therefore, allows for molecular diagnosis without a prior hypothesis. However, the expertise and costs involved in analysing the very big data sets are unclear at this stage and the complexity required for this type of data analysis means that WGS remains only in the realm of a select few groups and not available to routine healthcare providers.
PRECONCEPTION CARRIER SCREENING
Single-Gene Carrier Screening Programmes Fail to Identify Most Carrier Couples
Carrier screening can be considered at various life stages, although the best time to offer screening is during the preconception period.65 Preconception carrier screening is used to identify couples at elevated risk of conceiving a child with a genetic disease and enables them to consider alternative reproductive options in certain populations.66 It can be used before pregnancy (preconception screening) or during pregnancy (prenatal screening). In the past, carrier screening programmes have focussed on a small number of common recessive disorders.65 This ethnicity-based screening approach was adopted partly because of the cost of screening for each condition.67
Carrier screening is primarily offered to individuals who have no knowledge of any disease in their family, as carriers are phenotypically healthy individuals. This approach is used for population screening campaigns in the general healthy population. Using this approach, only a minority of carrier couples are identified because the majority of affected children are born to couples with no known family history, and only a minority of relatives in high-risk families request carrier testing. A disadvantage of ancestry-based screening is that diseases are not limited to specific groups and it is difficult to assess who is at risk because of multi-ethnic backgrounds.65 The additional time and cost burdens of single-gene tests mean that screening for multiple conditions can be prohibitively expensive. Patients who might benefit from screening may not have access to the technology if they are outside of the identified high-risk groups.
Recent developments in genetic technologies have changed the way that carrier testing is conducted. Expanded carrier screening (ECS) panels have replaced multiple single-gene tests with a single assay. Using ECS, it is possible to assess hundreds of mutations associated with genetic disease, simultaneously screening for many different diseases and detecting a much larger set of sequence variants.65,68 Commercially available carrier screening tests include Applied Biosystems™ CarrierScan™ Assay (Thermo Fisher Scientific), Counsyl Foresight™ Carrier Screen (Myriad Genetics, Salt Lake City, Utah, USA), QHerit™ Expanded Carrier Screen (Quest Diagnostics, Secaucus, New Jersey, USA), and Inheritest® Carrier Screening (LabCorp®, Burlington, North Carolina, USA) (Table 1). Responsible implementation of ECS raises many technical, ethical, legal, and social questions, and there remains debate around which diseases should be tested, optimal pre and post-test counselling, and the potential impact on clinical and public health outcomes.65,69
FUTURE OUTLOOK
Innovations in genomic medicine offer exciting new opportunities in RH but also present new challenges. As advances in technology provide access to ever greater amounts of genetic information, patients and healthcare providers (HCP) are faced with ethical and practical considerations. Ease of data analysis and interpretation followed by genetic counselling will become increasingly important as patients and HCP consider their options in light of improved genetic knowledge.65,70
A number of factors have been cited as barriers to the uptake of new genetic technologies, including a lack of adequate education among HCP and patients, prohibitive costs or lack of reimbursement, lack of counselling and support services, and lack of clear guidelines and policies for their use.65 Consequently, patient access to new genetic technologies varies greatly among different communities and countries.
CONCLUSION
Advances in molecular technologies have changed current practices in RH. Traditional microscope-based technologies are being replaced by molecular genetic techniques that are more efficient, more flexible, have a higher throughput and resolution, are potentially non-invasive, and greatly increase the diagnostic yield. The use of NGS in preimplantation genetic testing has dramatically increased our ability to detect aneuploid embryos and other genetic abnormalities and has substantially improved embryo selection for IVF.71 ERA is replacing histological assessments of endometrial receptivity, greatly improving pregnancy rates in IVF patients with RIF of endometrial origin. CMA detects smaller structural chromosome abnormalities and single-exon level deletions and duplications in addition to CNV in prenatal and postnatal samples. NGS has also revolutionised the detection of point mutations, enabling the parallel testing of many or all genes at the same time, thus accelerating the discovery of genetic causes of intellectual disability, birth defects, and rare genetic and genomic disorders. More recently, these methods have also been used with cfDNA in the preimplantation and prenatal diagnostics settings, avoiding the need for invasive procedures. Expanded carrier screening enables pan-ethnic screening regardless of a patient’s ethnicity, reducing the risk of stigmatising ethnic groups and without a significant increase in cost. The increased genetic information afforded by these technologies is accompanied by ethical and practical challenges for patients, HCP, and payers.








