Abstract
Ultrasound is one of the most ubiquitous imaging modalities in clinical practice. It is cheap, does not require ionising radiation, and can be performed at the bedside, making it the most commonly used imaging technique in pregnancy. Despite these advantages, it does have some drawbacks such as relatively low imaging quality, low contrast, and high variability. With these constraints, automating the interpretation of ultrasound images is challenging. However, successfully automated identification of structures within three-dimensional ultrasound volumes has the potential to revolutionise clinical practice. For example, a small placental volume in the first trimester is correlated to adverse outcome later in pregnancy. If the placenta could be segmented reliably and automatically from a static three-dimensional ultrasound volume, it would facilitate the use of its estimated volume, and other morphological metrics, as part of a screening test for increased risk of pregnancy complications, potentially improving clinical outcomes.
Recently, deep learning has emerged, achieving state-of-the-art performance in various research fields, notably medical image analysis involving classification, segmentation, object detection, and tracking tasks. Due to its increased performance with large datasets, deep learning has garnered great interest relating to medical imaging applications. In this review, the authors present an overview of deep learning methods applied to ultrasound in pregnancy, introducing their architectures and analysing strategies. Some common problems are presented and some perspectives into potential future research are provided.
INTRODUCTION
In medical imaging, the most commonly employed deep learning methods are convolutional neural networks (CNN).1-8 Compared to classical machine learning algorithms, CNN have enabled the development of numerous solutions not previously achievable because they do not need a human operator to identify an initial set of features: they can find relevant features within the data itself. In many cases, CNN are better able to identify features than the human eye. CNN have some disadvantages however: they need large amounts of data to automatically find the right features and processing large datasets is both computationally costly and takes time. Fortunately, training time can be reduced significantly if parallel architectures are used (e.g., by using graphics cards).
In medical imaging, deep learning is increasingly used for tasks such as automated lesion detection, segmentation, and registration to assist clinicians in disease diagnosis and surgical planning. Deep learning techniques have the potential to create new screening tools, predict diseases, improve diagnostic accuracy, and accelerate clinical tasks, whilst also reducing costs and human error.9-17 For example, automated lesion segmentation tools usually run in a few seconds, much faster than human operators, and often provide more reproducible results.
Ultrasound is the most commonly used medical imaging modality for diagnosis and screening in clinical practice.18 It presents many advantages over other modalities such as X-ray, magnetic resonance imaging (MRI), and computed tomography (CT) because it does not use ionising radiation, is portable, and is relatively cheap.19 However, ultrasound has its disadvantages. It often has relatively low imaging quality, is prone to artefacts, is highly dependent on operator experience, and has high inter- and intra-observer variability across different manufacturers’ machines.10 Nonetheless, its safety profile, noninvasive nature, and convenience makes it the primary imaging modality for fetal assessment in pregnancy.20 This includes early pregnancy dating, screening for fetal structural abnormalities, and the estimation of fetal weight and growth velocity.21 Although two-dimensional (2D) ultrasound is most commonly used for pregnancy evaluation due to its wide availability and high resolution, most machines also have three-dimensional (3D) probes and software, which have been successfully employed to detect fetal structural abnormalities.22
Ultrasound has a number of limitations when it comes to intrauterine scanning, including small field-of-view, poor image quality under certain conditions (e.g., reduced amniotic fluid), limited soft-tissue acoustic contrast, and beam attenuation caused by adipose tissue.22 Furthermore, fetal position, gestational age-induced effects (poor visualisation, skull ossification), and fetal tissue definition can also affect the assessment.20 As a result, a high level of expertise is essential to ensure adequate image acquisition and appropriate clinical diagnostic performance. Thus, ultrasound examination results are highly dependent on the training, experience, and skill of the sonographer.23
A study of the prenatal detection of malformations using ultrasound images demonstrated that the performance sensitivity ranged from 27.5% to 96.0% among different medical institutions.24 Even when undertaken correctly by an expert, manual interrogation of ultrasound images is still time-consuming and this limits its use as a population-based screening tool.
To address these challenges, automated image analysis tools have been developed which are able to provide faster, more accurate, and less subjective ultrasound markers for a variety of diagnoses. In this paper, the authors review some of the most recent developments in deep learning which have been applied to ultrasound in pregnancy.
DEEP LEARNING APPLICATIONS IN PREGNANCY ULTRASOUND
Deep learning techniques have been used for ultrasound image analysis in pregnancy to address such tasks as classification, object detection, and tissue segmentation. This review covers applications in pregnancy. The reviewed papers were identified with a broad free-text search on the most commonly used medical databases (PubMed, Google Scholar etc.). The search was augmented by reviewing the references in the identified papers. The resulting papers were assessed by the authors and filtered for perceived novelty, impact in the field, and published date (2017-2020). Table 1 lists the literature reviewed in this section.
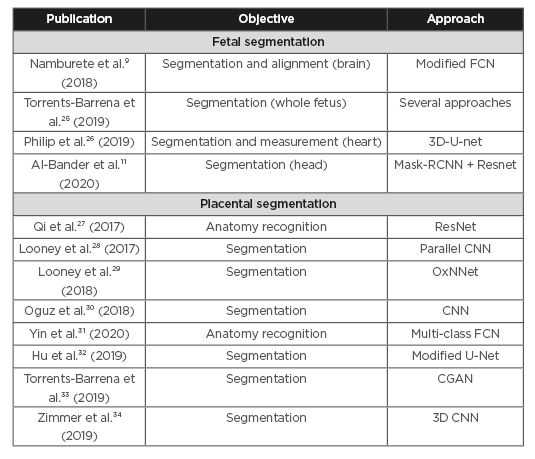
Table 1: Literature reviewed.
CGAN: conditional generative adversarial network; CNN: convolutional neural networks; FCN: fully convolutional neural network; RCNN: Regional Convolutional Neural Network.
Fetal Segmentation
Ultrasound is the imaging modality most commonly used in routine obstetric examination.
Fetal segmentation and volumetric measurement have been explored for many applications, including assessment of the fetal health, calculation of gestational age, and growth velocity. Ultrasound is also used for structural and functional assessment of the fetal heart, head biometrics, brain development, and cerebral abnormalities. This antenatal assessment allows clinicians to make an early diagnosis of many conditions, facilitating parental choice and enabling appropriate planning for the rest of the pregnancy including early delivery.
Currently, fetal segmentation and volumetric measurement still rely on manual or semi-automated methods, which are time-consuming and subject to inter-observer variability.11 Effective fully automated segmentation is required to address these issues. Recent developments to facilitate automated fetal segmentation from 3D ultrasound are presented below:
Namburete et al.9 developed a methodology to address the challenge of aligning 3D ultrasound images of the fetal brain to form the basis of automated analysis of brain maturation. A multi-task fully convolutional neural network (FCNN) was used to localise the 3D fetal brain, segment structures, and then align them to a referential co-ordinate system. The network was optimised by simultaneously learning features shared within the input data pertaining to the correlated tasks, and later branching out into task-specific output streams.
The proposed model was trained on a dataset of 599 volumes with a gestational age ranging from 18 to 34 weeks, and then evaluated on a clinical dataset consisting of 140 volumes presenting both healthy and growth-restricted fetuses from different ethnic and geographical groups. The automatically co-aligned volumes showed a good correlation between fetal anatomies.
Torrents-Barrena et al.25 proposed a radiomics-based method to segment different fetal tissues from MRI and 3D ultrasound. This is the first time that radiomics (the high-throughput extraction of large numbers of image features from radiographic images35) has been used for segmentation purposes. First, handcrafted radiomic features were extracted to characterise the uterus, placenta, umbilical cord, fetal lungs, and brain. Then the radiomics for each anatomical target were optimised using both K-best and Sequential Forward Feature Selection techniques. Finally, a Support Vector Machine with instance balancing was adopted for accurate segmentation using these features as its input. In addition, several state-of-the-art deep learning-based segmentation approaches were studied and validated on a set of 60 axial MRI and 3D ultrasound images from pathological and clinical cases. Their results demonstrated that a combination of 10 selected radiomic features led to the highest tissue segmentation performance.
Philip et al.26 proposed a 3D U-Net based fully automated method to segment the fetal annulus (base of the heart valves). The aim of this was to build a tool to help fetal medicine experts with assessment of fetal cardiac function. The method was trained and tested on 250 cases (at different points in the cardiac cycle to ensure that the technique was valid). This provided automated measurements of the excursion of the mitral and tricuspid valve annular planes in form of TAPSE/MAPSE (TAPSE: tricuspid annular plane systolic excursion; MAPSE: mitral annular plane systolic excursion). This demonstrated the feasibility of using this technique for automated segmentation of the fetal annulus.
Al-Bander et al.11 introduced a deep learning-based method to segment the fetal head in ultrasound images. The fetal head boundary was detected by incorporating an object localisation scheme into the segmentation, achieved by combining a Mask R-CNN (Regional Convolutional Neural Network) with a FCNN. The proposed model was trained on 999 3D ultrasound images and tested on 335 images captured from 551 pregnant women with a gestational age ranging between 12 and 20 weeks. Finally, an ellipse was fitted to the contour of the detected fetal head using the least-squares fitting algorithm.3 Figure 1 illustrates the examples of fetal head segmentation.
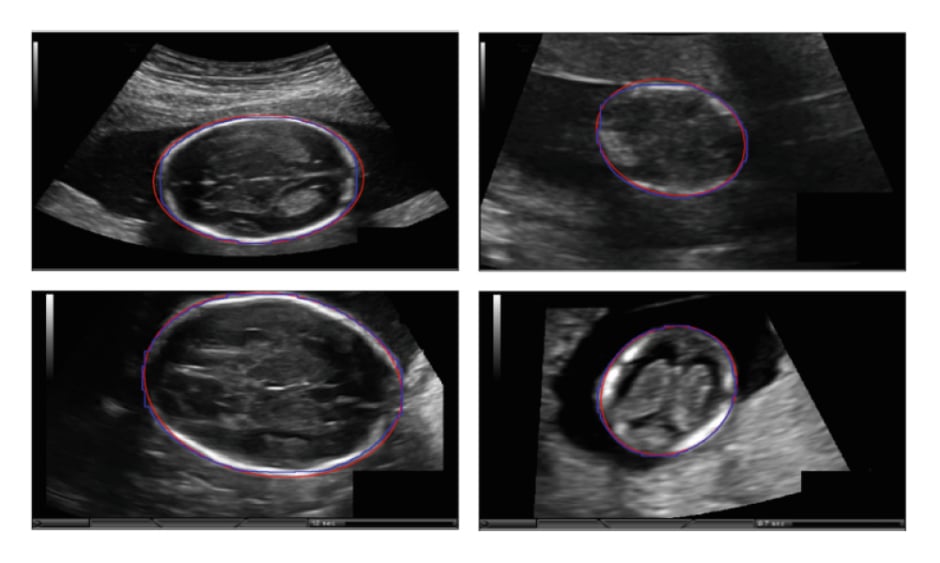
Figure 1: Examples of fetal head segmentation showing the ellipse fitted results on the two-dimensional ultrasound sections.
Manual annotation (blue); automated segmentation (red).
Adapted from Al-Bander et al.11
Placental Segmentation
The placenta is an essential organ which plays a vital role in the healthy growth and development of the fetus. It permits the exchange of respiratory gases, nutrients, and waste between mother and fetus. It also synthesises many substances that maintain the pregnancy, including oestrogen, progesterone, cytokines, and growth factors. Furthermore, the placenta also functions as a barrier, protecting the fetus against pathogens and drugs.37
Abnormal placental function affects the development of the fetus and causes obstetric complications such as pre-eclampsia. Placental insufficiency is associated with adverse pregnancy outcomes including fetal growth restriction, caused by insufficient transport of nutrients and oxygen through the placenta.38 A good indicator of future placental function is the size of the placenta in early pregnancy. The placental volume as early as 11 to 13 weeks’ gestation has long been known to correlate with birth weight at term.39 Poor vascularity of the first-trimester placenta also increases the risk of developing pre-eclampsia later in pregnancy.40
Reliable placental segmentation is the basis of further measurement and analysis which has the ability to predict adverse outcomes. However, full automation of this is a challenging task due to the heterogeneity of ultrasound images, indistinct boundaries, and the placenta’s variable shape and position. Manual segmentation is relatively accurate but is extremely time-consuming. Semi-automated image analysis tools are faster but are still time-consuming and typically require the operator to manually identify the placenta within the image. An accurate and fully automated technique for placental segmentation that provides measurements such as placental volume and vascularity would permit population-based screening for pregnancies at risk of adverse outcomes. Figure 2 illustrates an example of placenta segmentation.
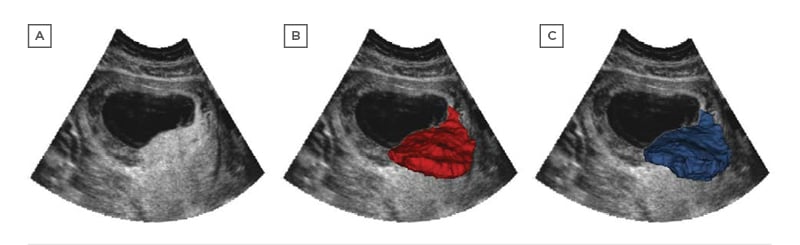
Figure 2: Placenta segmentation of first-trimester pregnancy: A) two-dimensional B-mode plane; B) semi-automated Random Walker result; C) OxNNet prediction result.
Adapted from Looney et al.29
Qi et al.27 proposed a weakly supervised CNN for anatomy recognition in 2D placental ultrasound images. This was the first successful attempt at multi-structure detection in placental ultrasound images.
The CNN was designed to learn discriminative features in Class Activation Maps (one for each class), which are generated by applying Global Average Pooling in the last hidden layer. An image dataset of 10,808 image patches from 60 placental ultrasound volumes were used to evaluate the proposed method. Experimental results demonstrated that the proposed method achieved high recognition accuracy, and could localise complex anatomical structures around the placenta.
Looney et al.28 used a CNN named DeepMedic41 to automate segmentation of placenta in 3D ultrasound. This was the first attempt to segment 3D placental ultrasound using a CNN. Their database contained 300 3D ultrasound volumes from the first trimester. The placenta was segmented in a semi-automated manner using the Random Walker method,42 to provide a ‘ground truth’ dataset. The results of the DeepMedic CNN were compared against semi-automated segmentation, achieving median Dice similarity coefficient (DSC) of 0.73 (first quartile, third quartile: 0.66, 0.76) and median Hausdorff distance of 27 mm (first quartile, third quartile: 18 mm, 36 mm).
Looney et al.29 then presented a new 3D FCNN named OxNNet. This was based on the 2D U-net architecture to fully automate segmentation of the placenta in 3D ultrasound volumes. A large dataset, composed of 2,393 first trimester 3D ultrasound volumes, was used for training and testing purposes. The ground truth dataset was generated using the semi-automated Random Walker method42 (initially seeded by three expert operators). The OxNNet FCNN obtained placental segmentation with state-of-the-art accuracy (median DSC of 0.84, interquartile range 0.09). They also demonstrated that increasing the size of the training set improves the performance of the FCNN. In addition, the placental volumes segmented by OxNNet were correlated with birth weight to predict small-for-gestational-age babies, showing almost identical clinical conclusions to those produced by the validated semi-automated tools.
Oguz et al.30 combined a CNN with multi-atlas joint label fusion and Random Forest algorithms for fully automated placental segmentation. A dataset of 47 ultrasound volumes from the first trimester was pre-processed by data augmentation. The resulting dataset was used to train a 2D CNN to generate a first 3D prediction. This was used to build a multi-atlas joint label fusion algorithm, generating a second prediction. These two predictions were fused together using a Random Forest algorithm, enhancing overall performance. A four-fold cross-validation was performed and the proposed method reportedly achieved a mean Dice coefficient of 0.863 (±0.053) for the test folds.
Yin et al.31 proposed a fully automated method combining deep learning and image processing techniques to extract the vasculature of the placental bed from 3D power Doppler ultrasound scans and estimate its perfusion. A multi-class FCNN was applied to separate the placenta, amniotic fluid, and fetus from the 3D ultrasound volume to provide accurate localisation of the utero-placental interface (UPI) where the maternal blood enters the placenta from the uterus. A transfer learning technique was applied to initialise the model using parameters optimised by a single-class model²⁹ trained on 1,200 labelled placental volumes. The vasculature was segmented by a region growing algorithm from the 3D power Doppler signal. Based on the representative vessels at a certain distance from the UPI, the perfusion of placental bed was estimated using a validated technique known as FMBV (fractional moving blood volume).43
Hu et al.32 proposed a FCNN based on the U-net architecture for 2D placental ultrasound segmentation. The U-net had a novel convolutional layer weighted by automated acoustic shadow detection, which helped to recognise ultrasound artefacts. The dataset used for evaluation contained 1,364 fetal ultrasound images acquired from 247 patients over 47 months. The dataset was diverse because the image data was acquired from different machines operated by different specialists and presented scanning of fetuses at different gestational ages. The proposed method was first applied across the entire dataset and then over a subset of images containing acoustic shadows. In both cases, the acoustic shadow detection scheme was proven to be able to improve segmentation accuracy.
Torrents-Barrena et al.33 proposed the first fully automated framework to segment both the placenta and the fetoplacental vasculature in 3D ultrasound, demonstrating that ultrasound enables the assessment of twin-to-twin transfusion syndrome by providing placental vessel mapping. A conditional Generative Adversarial Network was adopted to identify the placenta, and a Modified Spatial Kernelized Fuzzy C-Means combined with Markov Random Fields was used to extract the vasculature. The method was applied on a dataset of 61 ultrasound volumes, which was heterogeneous due to different placenta positions, in singleton or twin pregnancies of 15 to 38 weeks’ gestation. The results achieved a mean Dice coefficient of 0.75±0.12 for the placenta and 0.70±0.14 for its vessels on images that had been pre-processed by down-sampling and cropping.
Zimmer et al.34 focussed on the placenta at late gestational age. Ultrasound scans are typically useful only in the early stages of pregnancy because a limited field of view only permits the complete capture of small placentas. To overcome this, a multi-probe system was used to acquire different fields of view and then combine them with a voxel-wise fusion algorithm to obtain a fused ultrasound volume capturing the whole placenta. The dataset used for evaluation was composed of 127 single 4D (3D+time) ultrasound volumes from 30 patients covering different parts of the placenta. In total, 42 fused volumes were derived from these simple volumes which extended the field of view. Both the simple and fused volumes were used for evaluation of their 3D CNN based automated segmentation. The best results of placental volume segmentation were comparable to corresponding volumes extracted from MRI, achieving Dice coefficient of 0.81±0.05.
DISCUSSION
The number of applications for deep learning in pregnancy ultrasound has increased rapidly over the last few years and they are beginning to show very promising results. Along with new advances in deep learning methods, new ultrasound applications are being developed to improve computer-aided diagnosis and enable the development of automated screening tools for pregnancy.
A number of deep learning algorithms have been presented in this review, showing novel approaches, state-of-the-art results, and pioneering applications that have contributed so far to the pregnancy ultrasound analysis. Some methods rely on sophisticated hybrid approaches, combining different machine learning or image analysis techniques, whilst others rely on smart manipulation of the dataset such as fusing volumes or applying data augmentation. Large quality-controlled datasets are enabling single deep learning algorithms to be successfully developed still. However, it’s not currently possible to compare these methods directly, even if designed for the same task, because they all use different datasets and measurements.
The technological advances in medical equipment and image acquisition protocols allow better data acquisition to enhance the trained models. The size and availability of quality-controlled ground-truth datasets remain significant issues to be addressed. The performance of deep learning methods usually depends on the number of samples. Most of the presented methods cannot be independently evaluated because their datasets are small and not widely available. In addition, models trained on one dataset might fail on another generated by a different manufacturer’s machine. Large, publicly available, and appropriately quality-controlled ultrasound datasets are needed to compare different deep learning methods and achieve robust performance in real world scenarios.
There is also an urgent need to implement deep learning methods to solve relevant clinical problems. Very few papers translate the simple application of algorithms to a broader, practical solution that could be widely used in clinical practice. The practical implementation of deep learning methods and assessment of the correlation between automated results and clinical outcomes should be a focus of future research.
CONCLUSION
The field of deep learning in pregnancy ultrasound is still developing. Lack of sufficient high-quality data and practical clinical solutions are some of the key barriers. In addition, the newest deep learning methods tend to be applied first to other more homogeneous medical imaging modalities such as CT or MRI. Therefore, there is a need for researchers to collaborate across modalities to transfer existing deep learning algorithms to the field of pregnancy ultrasound to achieve better performance and create new applications in the future.







