BACKGROUND AND AIMS
Gestational diabetes mellitus (GDM), defined as hyperglycaemia during pregnancy, is associated with an increased future offspring risk of insulin resistance and obesity later in life,1 suggesting that hyperglycaemia exposure during pregnancy could have an adverse effect on the offspring. This could be explained by an epigenetic mechanism in response to GDM, however, this link remains inconclusive. Therefore, the authors sought to perform the largest epigenome-wide association study (EWAS) using the Finnish Gestational Diabetes (FinnGeDi) prospective multicentre cohort2 to investigate epigenetic changes associated with GDM exposure during pregnancy in both mothers and offspring.
MATERIALS AND METHODS
The authors designed a case-control study using a total of 536 offspring–mother pairs, of which 55% were exposed to GDM. DNA extracted from whole blood for the mothers and from cord blood for the offspring was subjected to methylation analysis using the Infinium MethylationEPIC (850K) arrays (Illumina, San Diego, California, USA). The authors performed three EWAS, adjusted for age, BMI, maternal weight gain, and cellular composition (blood) for mothers, and adjusted for sex, gestational weight, gestational week, and cellular composition (cord blood) for the offspring. EWAS was performed to identify differentially methylated sites associated with mothers and offspring, separately; shared methylation sites between mothers and offspring; and offspring-specific effects, adjusted for maternal methylation, to account for maternal methylation status.
RESULTS
The present study did not identify any false discovery rate (FDR) significant sites associated with mothers or offspring, separately, nor shared sites between mothers and offspring. For the offspring-specific effects, the authors adjusted for the maternal methylome to account for the impact of the maternal environment. The authors identified a single FDR-significant site associated with GDM exposure: a hypomethylation at the cg22790973 probe, located upstream of the transcription start site of the TFCP2 gene.
Furthermore, the study also included an interaction term in the model between GDM exposure and maternal methylation (i.e., GDM exposure in the context of the maternal methylation status) and identified a further seven CpG sites, of which the most significant was the same cg22790973 probe (TFCP2), in addition to cg03456133, cg24440941 (H3C6), cg20002843 (LOC127841), cg19107264, cg11493553 (UBE3C) and cg17065901 (FAM13A), and cg23355087 (DLGAP2) (Figure 1). Of relevance, UBE3C and FAM13A are reported susceptibility genes for Type 2 diabetes and BMI,3,4 whereas the DLGAP2 gene was associated with insulin sensitivity during pregnancy.5
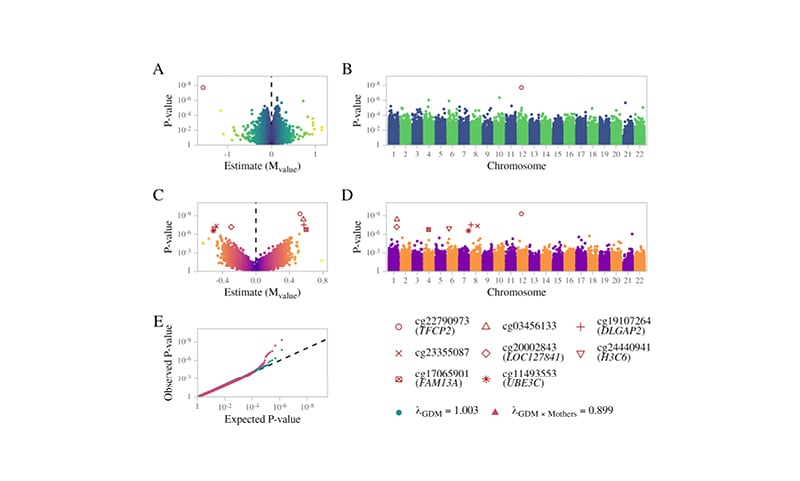
Figure 1: Summary results for EWAS associations.
A) Volcano plot and B) Manhattan plot for offspring differentially methylated sites associated with GDM exposure. C) Volcano plot and D) Manhattan plot of the GDM exposure interaction effect. E) Probability–probability plot of the GDM exposure main effect (green) and interaction term (red) on the methylation of the offspring. The black line illustrates the expected distribution.
GDM: gestational diabetes mellitus.
CONCLUSION
In conclusion, the authors present a comprehensive study investigating the epigenetic associations in response to GDM exposure in mother–offspring pairs. The study data do not support robust epigenetic associations for mothers and offspring exposed to GDM during pregnancy; however, in terms of offspring-specific effects, the maternal environment may have a moderating effect. Therefore, the authors identified a novel perspective in maternal transmission, determined by not only GDM exposure, but also other factors, such as maternal epigenetic status, that establish epigenetic signatures in offspring.