Meeting Summary
Use of bimekizumab, a monoclonal immunoglobulin G1 (IgG1) antibody that selectively inhibits the cytokine interleukin-17F (IL-17F), in addition to IL-17A, has been shown to result in a rapid response and prolonged, high-level disease control in patients with moderate-to-severe plaque psoriasis. At the 2024 European Academy of Dermatology and Venereology (EADV) Congress, an oral presentation and a poster were presented related to furthering the understanding of the mechanisms that lead to the clinical response observed with bimekizumab. The oral presentation examined the effects of bimekizumab on subcomponents of the Psoriasis Area and Severity Index (PASI). Analyses showed ≥95% mean improvement in erythema, thickness, and scaling with bimekizumab by Week 12 of treatment, which was maintained to Week 48. Bulk transcriptomic analysis showed complete normalisation of gene signatures associated with these subcomponents by Week 8, preceding clinically apparent skin clearance. A poster presented at the 2023 5th Inflammatory Skin Diseases (ISDS) focused on the effect of bimekizumab on tissue-resident memory T (TRM) cells, which have been associated with disease perpetuation during treatment and with psoriasis recurrence at the same location following treatment withdrawal. Bulk transcriptomic analyses showed normalisation of a TRM gene signature at Week 8 after only two doses of bimekizumab, as well as normalised expression of pro-survival factors that may be prolonging survival of pathogenic TRM cells and pathogenic IL-17A/F-secreting cells. These results may have implications for disease modification and help explain the long-term durability of response observed with bimekizumab. The results shown in the oral presentation and poster support the rationale for initiation of the BE UNIQUE study, the protocol for which was reported at EADV 2024. This ongoing, multicentre, Phase IIIb study is recruiting patients with moderate-to-severe plaque psoriasis, with or without concomitant active psoriatic arthritis, with a primary objective to assess change in composite gene expression score at Week 48. This study aims to further elucidate the molecular mechanisms underlying the rapid, high-level, and durable clinical responses observed with bimekizumab.
Introduction
The pro-inflammatory cytokines IL-17A and IL-17F are overexpressed in the thick, scaly, erythematous plaques that are the signature of skin manifestations in psoriasis.1-4 While IL-17A is the more potent of the two cytokines,5 IL-17F is more abundant in inflamed psoriatic tissue6 and chronic stimulation of IL-17-secreting cells, especially T helper 17 cells, preferentially leads to IL-17F production.7
Bimekizumab is a humanised monoclonal IgG1 antibody with unique dual specificity for IL-17A and IL-17F.8 Use of bimekizumab is associated with immune response suppression and can lead to rapid and prolonged, high-level control of moderate-to-severe plaque psoriasis. This was demonstrated in the Phase III clinical studies BE SURE (NCT03412747),9 BE VIVID (NCT03370133),10 and BE READY (NCT03410992),11 and their open-label extension study BE BRIGHT (NCT03598790).12 Among 989 patients randomised to receive subcutaneous bimekizumab in these studies, 62.7% achieved complete skin clearance at Week 16, measured as 100% improvement from baseline PASI score (PASI 100), and 87.5% achieved ≥90% improvement from baseline PASI score (PASI 90).12 Responses were durable, with 72.6% of bimekizumab-randomised patients who entered BE BRIGHT achieving PASI 100 after 4 years and 88.0% achieving PASI 90.13
Bimekizumab use has shown superior efficacy compared with placebo10,11 and with other biologics such as adalimumab,9 ustekinumab,10 and secukinumab.14 For example, in the BE RADIANT Phase IIIb trial, bimekizumab showed superiority to secukinumab at the primary endpoint, with 61.7% of the bimekizumab group (n=373) achieving PASI 100 at Week 16 compared with 48.9% of the secukinumab group (n=370; p<0.001).14 Here, findings from an oral presentation and two posters are highlighted to show the latest molecular understandings underpinning the observed clinical response with bimekizumab.
Rapid Normalisation of Molecular Signatures Associated with Psoriasis Area and Severity Index Subcomponents Following Bimekizumab Administration
PASI is the most widely used severity scoring system to assess psoriasis drug efficacy in the clinical setting. It consists of a composite score of three lesion-associated subcomponents (erythema, thickness, and scaling), weighted by plaque area and body region. The subcomponents are infrequently reported, but separate analysis should enable understanding of the contribution of each to the composite score and whether any subcomponent is obscured in the overall weighted composite score.15
In the findings presented in this oral presentation,16 PASI subcomponent data from the BE RADIANT trial was analysed. In this trial, participants were randomised to receive 320 mg bimekizumab every 4 weeks (Q4W) from Week 0 to Week 16, then either Q4W or Q8W until Week 48 (n=373), or 300 mg secukinumab weekly to Week 4 then Q4W (n=370).14
As can be seen from Figure 1, by Week 8 there was a mean reduction from baseline in lesion-associated erythema of 89.0%, in thickness of 92.1%, and in scaling of 93.3%. By Week 12, all three subcomponents showed ≥95% mean improvement, which was maintained to Year 1. As results were similar between the subcomponents, this suggests that analysis of any subcomponent alone is potentially reliable for evaluating clinical response to bimekizumab.16
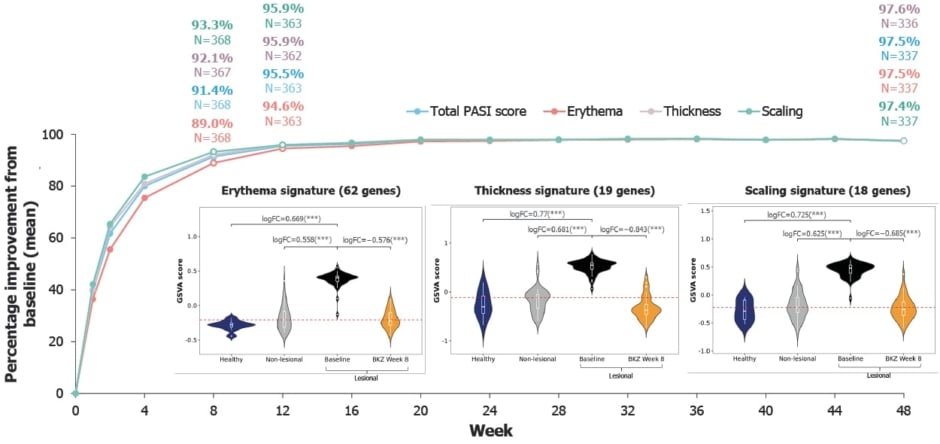
Figure 1: PASI mean percentage reduction from baseline across three subcomponents in the Phase IIIb BE RADIANT trial, and expression of subcomponent gene signatures in treated lesional tissue at Week 8 versus baseline non-lesional and lesional tissue, and healthy tissue in a Phase IIa trial.16
PASI subcomponent data are presented using observed cases for bimekizumab-randomised patients only (N=373) from the BE RADIANT Phase IIIb trial. Patients with a weighted score of 0 for a given PASI subcomponent at baseline were excluded from the analysis for that subcomponent. Molecular data is from a Phase IIa trial. For PASI subcomponent signature analysis, thickness was defined by the acanthosis gene set. Violin plots show expression of PASI subcomponent signatures, using GSVA to estimate gene set expression levels in healthy tissue (patients without psoriasis: blue), baseline non-lesional (clear skin in patients with psoriasis: grey), baseline lesional (patients with psoriasis: black), and treated lesional tissue at Week 8 (BKZ Week 8: orange). Wider sections of the violin plot indicate higher density of data at the respective y-axis value. White box plots show median and interquartile range normalised expression. Red horizontal lines correspond to median baseline expression in non-lesional tissue. LogFC and FDR-adjusted p-values were calculated using the limma moderated t-test. ***FDR<0.001.
BZK: bimekizumab; GSVA: Gene Set Variation Analysis; FC: fold change; PASI: Psoriasis Area and Severity Index.
To help understand the observed clinical response to bimekizumab, it is vital to elucidate the drug’s molecular effects. PASI subcomponent-linked gene sets were curated using two public gene ontologies: DisGeNET for erythema17 and the Human Phenotype Ontology for thickness (‘acanthosis’ in the ontology) and scaling.18 Gene sets of interest were then refined to include only the genes known to be dysregulated in psoriatic lesional versus non-lesional skin. These had been shown in the analysis of data from a Phase IIa clinical trial (NCT03025542)19 where 49 participants received two injections of 320 mg bimekizumab, 4 weeks apart, which led to rapid and profound normalisation of the psoriasis transcriptome by Week 8.19
Bulk RNA-sequencing data from this aforementioned Phase IIa trial19 was used to assess dysregulation of the psoriasis-specific PASI subcomponent-linked gene sets at baseline and following bimekizumab treatment.16 These data were generated from lesional and non-lesional skin biopsies previously collected at baseline and Week 8 and on tissue from healthy controls.19 Gene set level expression changes were assessed using gene set variation analysis and limma statistical methods.16
As shown in Figure 1, at baseline, signatures of all the PASI subcomponents were significantly upregulated compared with healthy and non-lesional tissue. At Week 8, following two doses of bimekizumab, complete normalisation of the subcomponent gene sets to non-lesional levels and beyond was observed: 95.7% median percentage improvement for erythema, 105.3% for thickness, and 104.5% for scaling.16
As bimekizumab selectively inhibits IL-17A and IL-17F,8 analysis was carried out to understand the relationship between the expression levels of these cytokines with the PASI subcomponent gene signatures (reported using Spearman correlation coefficients: R). Positive correlations were shown between baseline changes in IL17A and IL17F expression levels and mean gene expression changes of each curated gene set (R: 0.36−0.53; false discovery rate <0.05).15 This is consistent with the known direct effect of IL-17 on keratinocytes and indicates that both IL-17A and IL-17F are important in psoriatic disease.20
To further understand the molecular effects of bimekizumab on the PASI subcomponents, analysis was carried out on lesional psoriatic tissue with regard to selected markers associated with erythema (CXCL8),21 thickness (KRT16),22 and scaling (LORICRIN).23 Observation prior to treatment (utilising RNAscopeTM in situ hybridisation imaging) showed high expression of CXCL8 in epidermal layers associated with the presence of IL17F-expressing cells in lesional tissue, but only marginal expression in non-lesional tissue. Similar findings comparing lesional to non-lesional tissue were shown with regard to KRT16 in epidermal keratinocytes.16 Conversely, high levels of expression of LORICRIN, a barrier function gene,23 was shown in the granular layer of the epidermis in non-lesional tissue but was only present at low levels, if at all, in lesional tissue due to barrier loss associated with skin scaling.16
These observations were confirmed by analysis of these selected genes in the bulk RNA sequencing data from lesional and non-lesional baseline tissue reported here. Following bimekizumab treatment, by Week 8, similar analysis showed normalisation of these markers in the lesional tissue, with percentage improvements of 118.9% for CXCL8, 102.4% for KRT16, and 104.3% for LORICRIN.16
Overall, it was concluded that this first analysis of the effects of bimekizumab on PASI subcomponents revealed changes following administration at both clinical and molecular levels on all three subcomponents. These findings, the authors concluded, indicate clinical predictivity for sustainable skin clearance as early as 12 weeks, which is preceded by molecular resolution of disease.16
Understanding Bimekizumab’s Durable Clinical Response
TRM cells in lesional skin are implicated in psoriasis perpetuation during treatment and in location-specific lesion recurrence following treatment withdrawal.4 It is thus of interest to understand molecular mechanisms associated with TRM cells following treatment with bimekizumab, which is known to result in durable and continuous skin clearance.12
Firstly, in this poster presented at ISDS 2023, three independent single-cell RNA sequencing datasets6,24,25 (previously obtained from psoriatic lesion biopsies) were reprocessed in a uniform manner. Results were then used to assess gene expression in specific cell types, including TRM cells and those that express IL17A and IL17F.26
Analysis of each single-cell dataset showed that approximately 5% of TRM cells in lesional psoriatic tissue expressed IL17A/F, with additional analysis suggesting that a considerable proportion of all IL17A/F expression may come from TRM cells. Analysis of these single-cell datasets also showed highly similar transcriptomes in T cells that express IL17A and IL17F, with Pearson correlation coefficients of 0.94−0.98 and p<0.0001.26
The IL-7 pathway is associated with cell survival through upregulation of anti-apoptotic genes such as BCL2 and BCLXL.27 Analysis of baseline lesional tissue single-cell datasets showed that the receptor for IL-7 (IL7R) was highly expressed on both IL-17A- and, particularly, IL-17F-producing T cells in two out of the three datasets. This observation led to the postulation that the upregulation of IL7R may increase the survival of pathogenic IL-17A/F-secreting cells.26 As a follow-up analysis, expression of IL7R was confirmed in TRM cells specifically, as was that of another T-cell pro-survival factor, the cytokine IL32,26 which has previously been shown to be increased in cells from psoriatic skin lesions.28
To further elucidate the role of TRM cells, the effect of bimekizumab on genes and gene signatures of interest was also evaluated using pre- and post-treatment bulk RNA sequencing data from the Phase IIa trial of patients with psoriasis discussed above (NCT03025542).19 Prior to treatment, gene set variation analysis of a TRM gene signature showed this to be significantly higher in lesional tissue when compared with tissue from healthy controls and with non-lesional tissue from patients with psoriasis. Normalisation of TRM gene signature was shown following two doses of bimekizumab (Figure 2A), with a median percentage improvement of 78.1% at Week 8.26 Seventeen participants in this study also received a third dose of bimekizumab at Week 16.19 In this group, median percentage improvement in TRM gene signature increased to 87.7% at Week 28 (Figure 2A).26
Prior to treatment, and compared with healthy and non-lesional tissue, upregulation of IL7R and IL32 expression was also shown in lesional tissue through bulk RNA sequencing. This was reversed following bimekizumab administration by Week 8, and furthermore by Week 28 (Figure 2B). Additionally observed was a baseline increase in an anti-apoptotic gene signature compared with healthy and non-lesional tissue, with bimekizumab administration leading to normalisation of this signature at Weeks 8 and 28 (median percentage improvement at Week 8 of 104.5%; Figure 2C).26
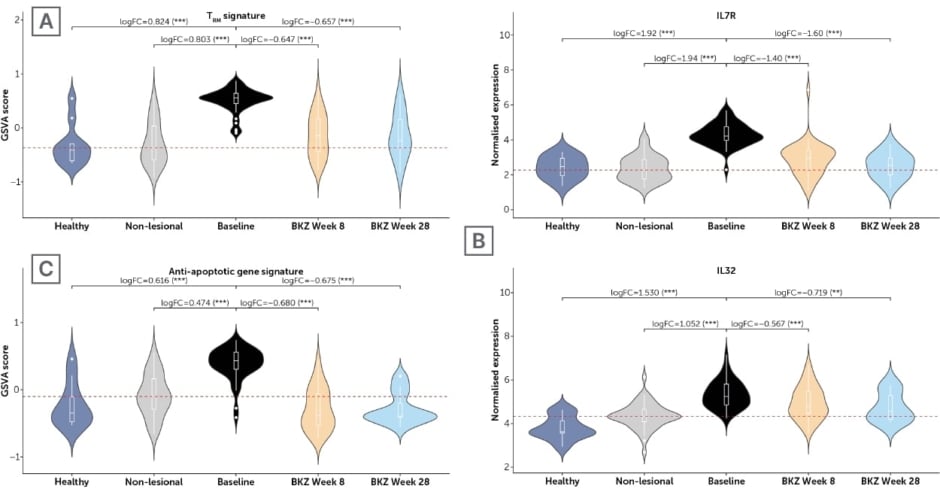
Figure 2: Expression of key genes and gene sets in baseline healthy, non-lesional and lesional tissue versus treated lesional tissue at Weeks 8/28 from a Phase IIa trial.26
Gene Set Variation Analysis was used to estimate gene set level of expression. Red horizontal lines correspond to the median baseline expression in non-lesional tissue. LogFC and false discover rate-adjusted p-values were calculated using the limma moderated t-test. ***false discovery rate<0.001; **false discovery rate<0.01. TRM signature: CD103, CD69, CD44; Anti-apoptotic gene signature: BCL2, BCL2L1, MCL1, BIRC5, CFLAR, BCL2A1, BIRC3, PEA 15.
BKZ: bimekizumab; GSVA: Gene Set Variation Analysis; IL: interleukin; TRM: tissue-resident memory T cells.
The authors concluded that ‘these mechanistic data from patient samples highlight the importance of IL-17F and IL-17A dual neutralisation in normalising both TRM cell biology and pro-survival factors’. Their findings may have implications for disease modification and for the maintenance of complete skin clearance during the treatment of psoriasis with bimekizumab.26
The BE UNIQUE Study
As discussed above, TRM cells are implicated in psoriasis4 and are found in the blood and joints of patients with psoriatic arthritis,29 a condition that occurs in around 15−20% of people with psoriasis.30,31 Findings showing rapid normalisation of the psoriatic skin transcriptome and TRM cell signatures following two injections of 320 mg bimekizumab every 4 weeks19,26 led to the development of the ongoing (as of October 2024) Phase IIIb, multicentre BE UNIQUE study (NCT06506916) presented at EADV 2024.32 This will enrol 80 adults with moderate-to-severe plaque psoriasis, defined as PASI ≥12, body surface area ≥10%, and Investigator’s Global Assessment ≥3. Of these 80, 40 participants will have concomitant, active, psoriatic arthritis, defined as disease meeting the Classification Criteria for Psoriatic Arthritis (≥1/68 tender joint count and ≥1/66 swollen joint count). Ten matching healthy controls will also be recruited.32
As can be seen from Figure 3, in Part 1 participants initially receive 320 mg bimekizumab Q4W to Week 16, then Q8W until Week 48. In Part 2 (Weeks 48–96), participants with PASI=0 (plus low disease activity for participants with psoriatic arthritis) will be randomised 1:1 to receive bimekizumab either Q8W or Q12W, and participants with PASI >0 and/or without low psoriatic arthritis disease activity will continue on a Q8W dosing schedule.32
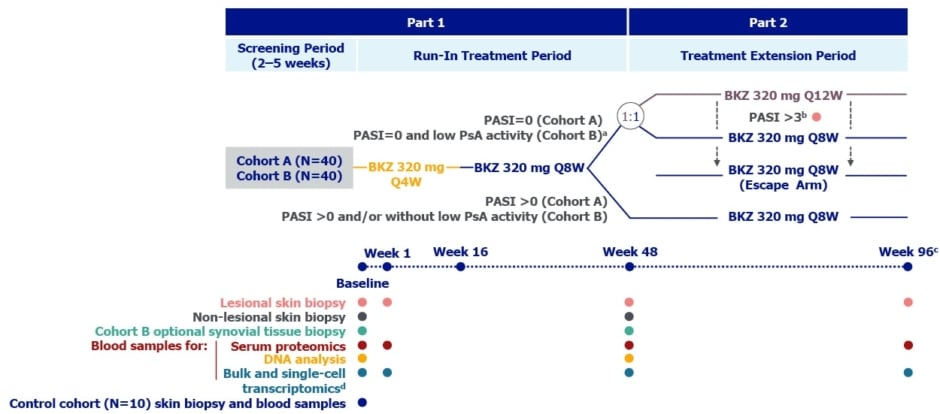
Figure 3: Study design for BE UNIQUE.32
*In addition to PASI=0, Cohort B patients must have low PsA disease activity at Week 48 (SJC ≤1 and no increase in concomitant medications for the treatment of PsA symptoms compared with baseline) to enter the Randomised Treatment Extension Period.
†Patients in Cohort A and Cohort B who have a PASI score >3 during the Randomised Treatment Extension Period will enter an Escape Treatment Period and receive bimekizumab Q8W to study end, undergoing additional assessments; a lesional skin biopsy will be taken at the visit the patient has a PASI score >3, instead of at Week 96.
‡ The safety follow-up visit will occur at least 12 weeks after the final dose and not before 4 weeks after the last skin biopsy.
§ Blood samples for single-cell transcriptomics will be collected from the subset of Cohort A and B patients from whom 6 mm skin biopsies are collected (3 mm or 6 mm skin biopsies will be possible) and from all Control Cohort participants. Blood samples for bulk transcriptomics will be collected from all study participants except the subset of Cohort A and B patients who undergo blood sampling for single-cell transcriptomics.
BKZ: bimekizumab; PASI: Psoriasis Area and Severity Index; PsA: psoriatic arthritis; Q4W: every 4 weeks; Q8W: every 8 weeks; Q12W: every 12 weeks; SJC: swollen joint count.
Lesional skin biopsies and blood samples for bulk and single-cell transcriptomics will be taken at baseline and at Week 1 (following the first bimekizumab injection), Week 48, and Week 96. Non-lesional skin biopsies will be taken at baseline and Week 48. For the cohort with psoriatic arthritis, biopsy of synovial tissue at baseline and Week 48 will be optional. Baseline skin biopsies and blood samples will be taken from a matching cohort of 10 healthy individuals.32
BE UNIQUE’s primary objective is assessment of change in composite gene expression score in the lesional skin biopsies using reverse transcription-polymerase chain reaction. This will utilise preselected genes based on known bimekizumab mechanisms of action and psoriatic disease pathways. The secondary objective is evaluation of the safety and tolerability of bimekizumab in these cohorts. Exploratory objectives include investigation of the effect of bimekizumab on skin and blood bulk, single cell, and spatial transcriptomics. Also assessed will be the clinical response to bimekizumab and systemic effects of bimekizumab on gene and protein expression in blood and serum, respectively.32
It is hoped, concluded the authors, ‘that BE UNIQUE will enable exploration of mechanisms underlying the rapid, high, durable clinical responses observed with bimekizumab treatment’, and will assess ‘whether durable clinical responses correlate with molecular, cellular, and transcriptomic changes in skin, blood, and joints of patients with psoriatic disease’.32
Conclusion
Together, these studies highlight how the utility of bimekizumab in the treatment of psoriasis may lie with the ability of this drug to impact underlying mechanisms associated with the development of this disease. Further investigation, in the BE UNIQUE study, will add to the understanding of the molecular and cellular changes brought about by treatment with bimekizumab.






