BACKGROUND
Volumetric segmentation of intrinsically heterogeneous abdominal tumours is essential for the diagnosis, follow-up, and treatment response evaluation of neuroblastomas.1 Manual segmentation of neuroblastic masses is a tedious and time-consuming task that hinders the radiologists’ workflow. Different studies explored semiautomatic segmentation algorithms in CT images, making use of mathematical morphology, fuzzy connectivity, and other image processing tools.2,3
As of today, a robust and generalisable solution does not exist for childhood neuroblastoma. In this study, the authors propose an automated segmentation method based on convolutional neural networks applied to children with neuroblastic tumours studied with multiple MRI sequences. The aim is to extract reproducible quantitative imaging biomarkers from these lesions for the prediction of relevant clinical outcomes.
MATERIALS AND METHODS
T1-weighted (T1W), T2-weighted (T2W), and diffusion-weighted (DW) MRI images at diagnosis were collected from 127 patients with neuroblastoma from different European hospitals in the scope of the H2020 PRIMAGE project.4 Images were subject to high variability in data acquisition due to different scanners, manufacturers, and protocols.
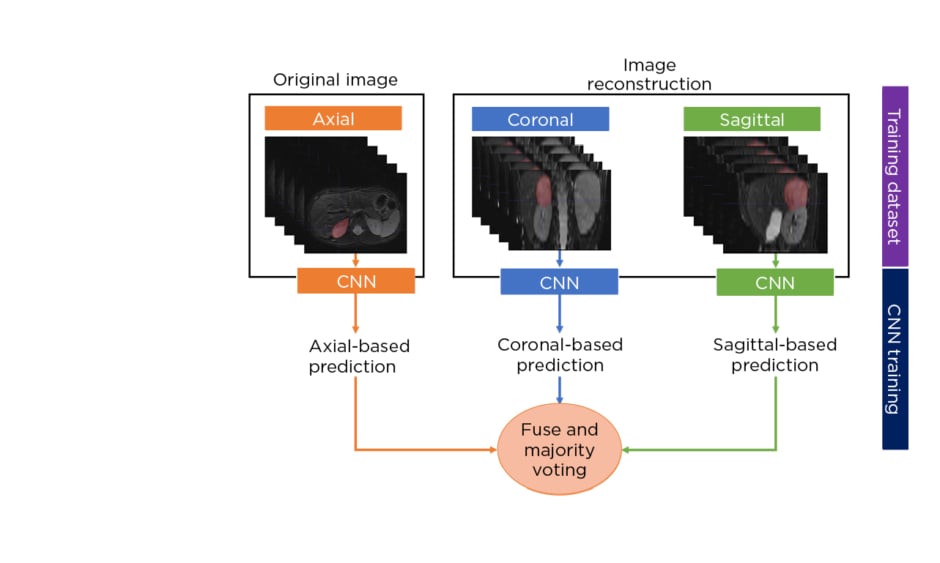
The authors developed a multisequence and multiplanar U-net.5 In order to justify the use of multisequence MRI, an experiment was performed with different inputs in a subset of randomly selected patients. For the multiplanar approach, the sagittal and coronal planes from the original axial MRI were reconstructed and fed into three separate neural networks. The segmentations from the three two-dimensional networks were fused using majority voting to generate the final 3D neuroblastoma segmentation (Figure 1[/h]). The networks were trained using the Adam optimiser with 300 epochs and a batch size of 100. The number of layers, number of convolutional feature maps, and learning rates were chosen based on hyperparameter tuning. In addition, a scheduler was used to set the learning rate in each epoch, starting from an initial value of 0.001 that reduced by a factor of 0.5 if the validation Dice plateaued for 20 epochs. The model was assessed by using a 5-fold cross-validation strategy and comparing the results with other state-of-the-art solutions.
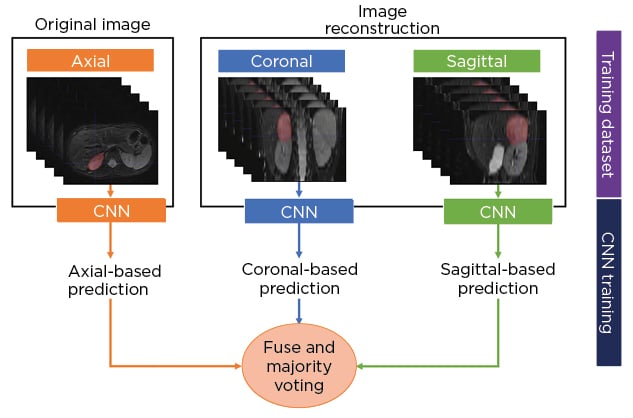
Figure 1: Schematic diagram of the multiplanar scheme used to develop a fully automated segmentation model of neuroblastic tumours.
CNN: convolutional neural network.
RESULTS
The Dice similarity coefficient values of the method using only T1W, T2W, DW, and a multisequence approach, having the 5-fold cross-validation as different inputs, were 0.732±0.064, 0.745±0.118, 0.786±0.077, and 0.841±0.038, respectively.
The average Dice similarity coefficient of the internal validation using the multisequence strategy was found to be 0.830, showing model robustness and stability across different sites and scanners.
CONCLUSION
The authors proposed a fully automated segmentation method of neuroblastic tumours based on convolutional neural networks and multisequence MRI with an accurate and stable performance. If further improved and externally validated the proposed method could be of use in clinical trials and oncologic practice for the management of neuroblastoma. Future work with a larger sample will be necessary to evaluate the generalisability of the model. In addition, the co-registration of T1W, T2W, and DW images is still particularly challenging in paediatric cancer. An artificial intelligence-based algorithm to overcome this limitation may benefit the segmentation process. ■