BACKGROUND AND AIMS
Patients with colorectal cancer (CRC) have limited systemic treatment options compared to other major cancer types.1 Tumour heterogeneity is a major cause of treatment failure.
MATERIALS AND METHODS
The authors have generated a living biobank of 208 patient-derived organoids (PDO) from liver metastases of 100 patients treated by hepatic resection for advanced CRC at Oslo University Hospital, Norway. The biobank includes multiple synchronous lesions (n=2–6) from 66 of the patients, and recurrent lesions sampled at hepatic re-resections of four patients. All PDOs have been screened for sensitivity to custom-made and clinically relevant drug libraries for CRC (n=40–47 drugs).2,3 Subsets of PDOs and corresponding tumour tissue samples have been analysed by multi-omics approaches.4 The study design is illustrated in Figure 1.
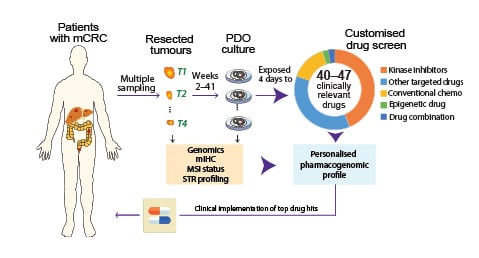
Figure 1: Study design.
Single or multiple metastases were resected from patients with advanced CRC and ex vivo PDOs established. Tumour organoids were seeded to pre-drugged plates, followed by cell viability measurements (3D CellTiter-Glo) after 96 hours. Drug Sensitivity Scores were calculated for each drug as a measure of reduced viability. In parallel, gene expression and sequencing of clinically relevant mutations were conducted for pharmacogenomic analyses. Short tandem repeat profiling was performed for all PDOs and their corresponding tissues for authentication.
Chemo: chemotherapy; CRC: colorectal cancer; mCRC: metastatic colorectal cancer; mIHC: multiplexed immunohistochemistry; MSI: microsatellite instability; PDO: patient-derived organoids; STR: short tandem repeat.
RESULTS
Metastatic lesions from individual patients showed only modest heterogeneity in drug sensitivities with distribution of mean Euclidean distances skewed towards lower intra-patient heterogeneity. Also, there was no correlation between the number of PDOs analysed per patient and their mean Euclidean distances. TP53 mutated PDOs were generally multidrug-resistant, and TP53 wild-type PDOs were sensitive to several chemotherapies, including 5-FU, SN-38, TAS-102, and gemcitabine. Furthermore, sensitivity to the PARP inhibitor talazoparib was significantly higher in TP53 wild-type PDOs, which supports the authors’ previous study suggesting wild-type TP53 activity as a mechanism of response to PARP inhibition in CRC cell lines.5 A TP53 mutated PDO harbouring an ATM mutation was hypersensitive to PARP inhibition, suggesting that other mechanisms of poly adenosine diphosphate-ribose polymerase inhibitor sensitivity are also involved. Among patients with a lack of sensitivity to standard-of-care drugs for CRC, 7% and 4% presented strong sensitivities towards conventional chemotherapies methotrexate and gemcitabine, respectively. Ex vivo drug vulnerabilities from the living biobank are currently being used as a reference basis for an interventional Phase II umbrella clinical study for patients with advanced CRC.
CONCLUSION
PDOs from multifocal liver metastases model pharmacogenomic heterogeneity of advanced CRCs and have strong potential for personalised oncology.








