BACKGROUND AND AIMS
Lung cancer (LC) remains the top cause of cancer-associated mortality worldwide, with a 10-year overall survival rate of only 5%.1 While most LCs are smoking-related, 25% of non-small cell LC (NSCLC) are diagnosed in patients with little or no smoking history.2 Fusions involving anaplastic lymphoma kinase (ALK) are the oncogenic driver in approximately 3–7% of NSCLC.3 While tyrosine kinase inhibitors (TKI) targeting the kinase domain of ALK have proven extremely effective, inevitably, resistance develops with limited effective treatment options.4
MATERIALS AND METHODS
The authors developed a precision medicine-based platform (PMP) to screen patient-derived material (PDM) directly from the operating room, with curated panels of drugs. PDM collected during clinically indicated procedures was plated in culture to generate patient-derived organoids, and screened with drugs curated to each tumour type. Patient-derived organoids are screened at therapeutically relevant doses, drawing from pharmacokinetic data for each drug, when available, in the authors’ previously published 3D assay.5 The authors have optimised an assay to rapidly screen for EML4-ALK fusions, and can perform next-generation DNA and RNA sequencing in real time (approximately 7 days) to integrate with drug screening results.
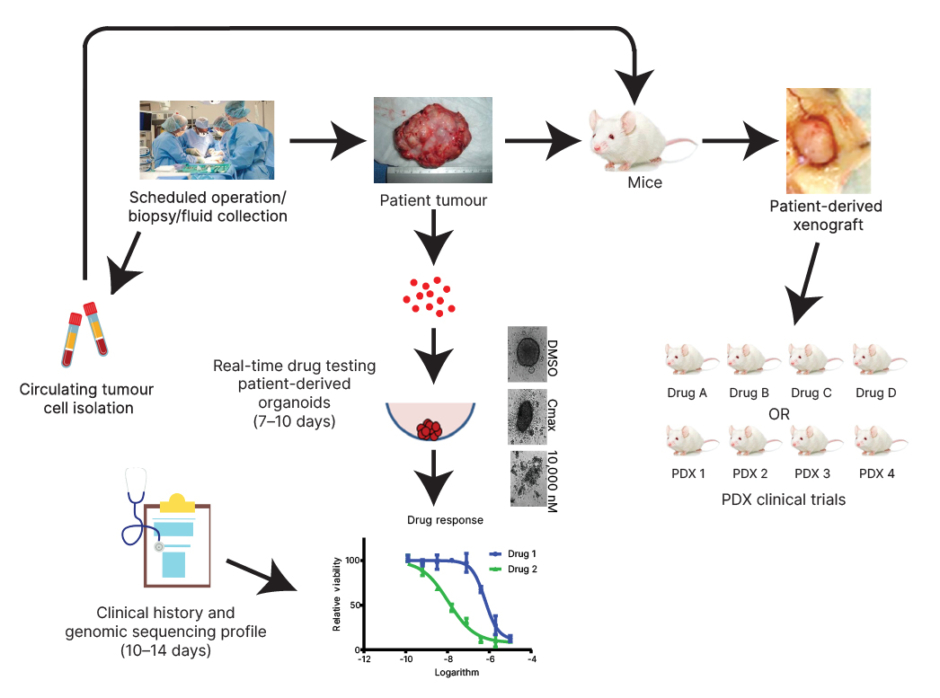
Figure 1: Schema for the precision medicine-based platform.
Patient tissue is brought into the lab following normally scheduled clinical procedures, and after obtaining informed consent. Material is utilised to obtain drug response information in real-time (7–10 days). Results are integrated with clinical history and molecular characterisation. When material is sufficient, renewable models like PDXs are developed for additional precision medicine studies.
Cmax: maximum plasma concentration; DMSO: dimethyl sulfoxide; PDX: patient-derived xenograft.
RESULTS
To date, the authors have screened 51 cases of NSCLC, including seven cases of ALK-positive NSCLC. Screening of EML4-ALK tumours, which have progressed to second- or higher line TKI, demonstrates sensitivity to earlier generation ALK TKIs, a known phenomenon in the literature. The authors’ results recapitulate known resistance in samples previously exposed to therapy, demonstrating a strong negative predictive value. Longitudinal assessment will be required to robustly assess positive predictive value. In one case of EML4-ALK NSCLC, the authors were able to collect PDM from two distinct regions (pleural effusion and paracentesis), and screen with the same panel of drugs, highlighting the reproducibility and consistency of their assay.
CONCLUSION
The authors’ PMP captures robust and reproducible results that are consistent with known clinical pathogenesis. Moving forward, they are collecting longitudinal data from enrolled patients in parallel with clinical trials, to demonstrate positive predictive value of their PMP. They additionally strive to demonstrate reproducibility to obtain Clinical Laboratory Improvement Amendments (CLIA) approval, and to deliver results to patients and physicians to help guide clinical care.







