Abstract
Background: There is an increasing incidence of life-threatening systemic mycoses, specifically fulminant infections by the Candida species in hospitalised patients and in those who are immunocompromised. Management of the limited number of antifungal drugs currently available requires the identification of infections containing drug-resistant isolates.
Objectives: The aim of this study was to identify the non-albicans Candida species as azole-resistant fungi, isolated from sputum and bronchoalveolar lavage specimens of hospitalised cases.
Methods: The subjects included hospital-acquired infection (HAI) cases, with a primary diagnosis using a direct microscopic examination, performed for the detection of probable fungi. The molecular tests of PCR-restriction fragment length polymorphism (RFLP) and real-time PCR were performed to confirm the identity and molecular typing of the Candida isolates. Antifungal susceptibility testing (AFST), by the Clinical and Laboratory Standards Institute (CLSI) broth microdilution (BMD) minimum inhibitory concentration (MIC) (M27-A2) method, was performed on the hospital-isolated
Candida species.
Results: During 24 months, from August 2014 to September 2016, a total of 198 samples were obtained from cases with proven HAI. The results of experimental studies on the specimens showed 93 (47%) positive cases for a fungal or bacterial infection, of which 54 (58%) had a fungal infection. It was hypothesised that all of the isolated organisms were causative agents of the HAI.
Conclusions: The results showed that the medium CHROMagar™ Candida is an accessible and easy-to-use method for the identification of infection, but not as accurate and reliable as PCR-RFLP and real-time PCR methods. Results also showed decreasing susceptibility to azoles (itraconazole in this study) of the Candida species.
BACKGROUND
The Candida species is the cause of several infections, including bloodstream infections and disseminated candidiasis. Despite recent advancements in the diagnosis and treatment of candidiasis, Candida is the fourth leading cause of bloodstream infection pathogens in the USA and seventh in Europe.1 Life-threatening systemic fungal infections have increased amongst immunocompromised patients, particularly fulminant infections caused by Candida spp. Patients immunocompromised as a result of HIV infection, allogenic haematologic stem cell transplantation, chemotherapy for acute leukaemia, or infant prematurity, are at risk of Candida infections.
Although Candida albicans is known as the most common cause of invasive candidiasis, there has been an increase in non-albicans infections, such as C. krusei, C. glabrata, C. lusitaniae, C. tropicalis, and C. parapsilosis. Some non-albicans Candida species, including C. auris, C. tropicalis, C. krusei, C. parapsilosis, and C. lusitaniae, are classified as notable pathogenic fungi that cause invasive candidiasis, mainly because of their innate resistance to antifungal drugs.2-6 Treating invasive fungal infections has been difficult because of the limited availability of antifungals, as well as the relative toxicity, drug interaction, and drug-resistance challenges.7 Systemic and invasive candidiasis can be treated with the limited number of available antifungal medications, such as polyenes, allylamines, azoles, and the newly produced echinocandin class of molecules.1 Adverse side effects, toxicity, and drug resistance in Candida species have been observed worldwide. Studies on the prevalence of Candida infections and antifungal susceptibility testing (AFST) can help to make decisions on clinical strategies. Studying the incidence of Candida infections, as well as AFST, is helpful in terms of planning clinical measures. Rapid identification of Candida spp. is also beneficial as early management of antifungal treatment can take place.1 Additionally, management of the limited number of antifungals currently available requires the identification of infections containing drug-resistant isolates, and the discovery of factors that promote the evolution of drug resistance.4
Objectives
The aim of this study was to identify the non-albicans Candida species from azole-resistant fungi, isolated from the sputum and bronchoalveolar lavage specimens of hospitalised cases. The performance of three rapid methods of identification at the species level was evaluated, and two susceptibility tests for the C. albicans clinical isolates were used.
METHODS
Subjects
Respiratory tract specimens were taken from patients with hospital-acquired infection (HAI) and infection symptoms occurring in the first 48 hours after arriving at the UMS University educational hospitals, in Urmia, Iran. Additionally, other clinical samples including pressure ulcers, skin abscess, vaginal discharge, and nail scrape were obtained from patients
during hospitalisation.
Culture and Identifications
The collected specimens were transported to the Medical Mycology Center, School of Medicine, UMS University. The primary diagnosis included direct microscopic examination (potassium hydroxide preparation [KOH]: 10%; Giemsa: 20% stained slides), which was performed for the detection of the possible fungi causing the HAI. This was followed by growing the cultures on sabouraud dextrose agar ([SDA]: 4%), Czapek yeast extract agar (CYA), corn meal agar, and CHROMagar™ Candida, for the identification of yeast isolates.8
Molecular Identification of Candida Species
First, DNA extraction was performed: molecular tests (PCR-restriction fragment length polymorphism [RFLP] and real-time PCR) were performed to confirm the identity and molecular typing of Candida isolates. DNA extraction of Candida cells was performed using phenol-chloroform and the glass beads manual method (making a lysis solution with 10 mM ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate, 100 mM sodium chloride, 100 mM Tris-HCl, and 1% TritonX-100 [Sigma-Aldrich, St Louis, Missouri, USA]).9
There are two important non-coding regions, internal transcribed spacers (ITS) 1 and 2, containing a variable subregion inside and a conservative subregion outside. The ITS fragments are located between the 18s and 28s rRNA genes of the Candida species. The variability of the ITS regions have proved useful in phylogenic studies of the Candida species. The target gene rRNA was amplified using ITS primers (ITS1: 5’-TCC GTA GGT GAA CCT GCG G-3’; ITS2: 5’-TCC TCC GCT TAT TGA TAT GC-3’). The PCR profile included: 5 µl of the DNA template; PCR buffer (20 mM Tris-HCl; pH 8.0); 50 mM potassium chloride; 0.1 mM each of the forward and reverse primers; and 1.5 U of Taq DNA polymerase in a total reaction volume of 50 µl. The reactions were performed in a Thermocycler model XL (Bioer Technology, Hangzhou, China). The PCR-RFLP, using the restriction enzyme MspI, was performed to create a differential pattern to identify the Candida species ([/hl]Table 1[/hl]; [/hl]Figure 1[/hl]).10 A real-time PCR for the confirmation of each RFLP-identified Candida isolate was carried out at the Molecular Research Center, EUMS, Esfahan, Iran.

Table 1: Clinical specimens collected from patients with Candida colonisation.
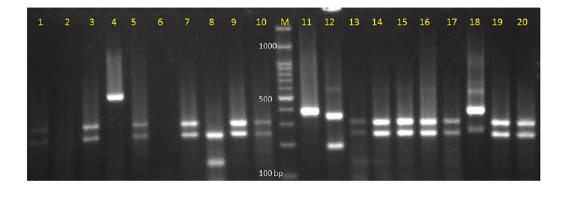
Figure 1: Electrophoretic picture of digestion with restriction enzyme MspI in the PCR-restriction fragment length polymorphism method.
Lanes: 1, 3, 5, 7, 9, 10, 13, 14, 15, 16, 19, 20 show C. albicans; lane 11: C. parapsilosis; lane 12: C. glabrata; lane 8:
C. tropicalis; lane 4: undigested PCR product; and lane M: 100 base pair DNA marker.
Susceptibility Test
AFST, by the Clinical and Laboratory Standards Institute (CLSI) broth microdilution (BMD) minimum inhibitory concentration (MIC) (M27-A2) method, was performed on hospital-isolated Candida species. The medium for testing antifungal agent constituted synthetic complete medium and RPMI-1640 (Sigma-Aldrich), supplemented with glutamine but without bicarbonate, as recommended. Supplementing medium to a final concentration of 20 g/L (2%) glucose has been shown to result in better growth of yeast isolates, without markedly altering the MIC of antifungal agents.10 Preparation of the stock drug solution included the antifungal drug itraconazole (Taj Pharmaceuticals Ltd., Birmingham, UK), prepared by weighing 16 mg on an analytical balance calibrated to 2 decimal places, as it was recommended that >16 mg (32 mg) of powder was used. Antifungal drug stock solution was prepared to a concentration 100 times the highest concentration to be tested (320 mg/mL).
Dimethyl sulfoxide (DMSO) was suitable for dissolving itraconazole, as well as other antifungal drugs (ketoconazole and flucytosine). The drug’s stock solutions were used, and the DMSO was prepared and sterilised beforehand.
For the yeast cell suspension, a 24-hour culture was prepared on 4% SDA and adjusted by a spectrophotometry system (wavelength: 530 nm; optical density: 75–77), ensuring that the yeast cell suspensions had equal concentration to the 0.5 McFarland standard.
Treatment
MIC testing of itraconazole was performed as per the procedure in the CLSI guidelines M27-A2,9-20 using RPMI-1640 broth, an inoculum of 0.5–2.5X103 colony-forming units (cfu)/mL, incubation at 35°C, and MIC endpoint criteria of prominent inhibition of growth.10 A serial dilution of the drug solution was made adding 100 µl of free bicarbonate RPMI-1640 into the first well (each lane of a 96-well plate), followed by a reduction in concentration (16.0000 to 0.0313 µg/mL). For the treatment, 100 µl of 1,000 times diluted yeast cells suspension was added to 10 wells, plus a negative control (without fungal cells) and a positive control (fungal cells only). After incubating for 24–48 hours at 35°C, turbidities of fungal growth were detected on a light screen. The MIC for the azole family were described as the lowest concentration of the drug that could reduce 50% of fungal growth. MIC of itraconazole for Candida cells were obtained by observing the first well without any growth.
RESULTS
Throughout the 24 months between August 2014 and September 2016, 198 clinical specimens were tested for fungal isolation. Bronchoalveolar lavage and sputum were the most frequent specimens with Candida isolation (as pathogens or colonising) ([/hl]Table 1[/hl]). Candida species, dominantly C. albicans, were obtained from 20/198 clinical specimens used in this study. Clinical cases were in the age range of 1–78 years. The identification results showed 93 (47.0%) patients with fungal or bacterial colonisation, and 25 (26.9%) had a fungal infection. It is thought that all of the isolated organisms were causing agents of infection in the tested cases. Among all isolated Candida species, C. albicans, C. dubliniensis, C. glabrata, and C. tropicalis were confirmed. These findings are based on the PCR-RFLP results confirmed by real-time PCR ([/hl]Table 2[/hl]). With the exception of C. dubliniensis, the results of molecular identifications were approximately similar in the two molecular methods. C. dubliniensis isolates in this study were identified by CHROMagar Candida and confirmed by real-time PCR. Additionally, a drug-resistant Candida isolate was identified as C. parapsilosis by PCR-RFLP and real-time PCR. Susceptibility testing (MIC) of Candida isolates results showed that 48% were resistant and 52% were sensitive to itraconazole. MIC results showed that tested C. parapsilosis were resistant against itraconazole (MIC: 0.125–0.250 μg/mL). Moreover, of the 23 tested samples of C. albicans/C. dubliniensis, 11 (47.8%) were resistant (MIC range: 0.125–0.250 μg/mL). Only one case of C. glabrata was sensitive to itraconazole with an MIC range of ≤0.125 μg/mL.
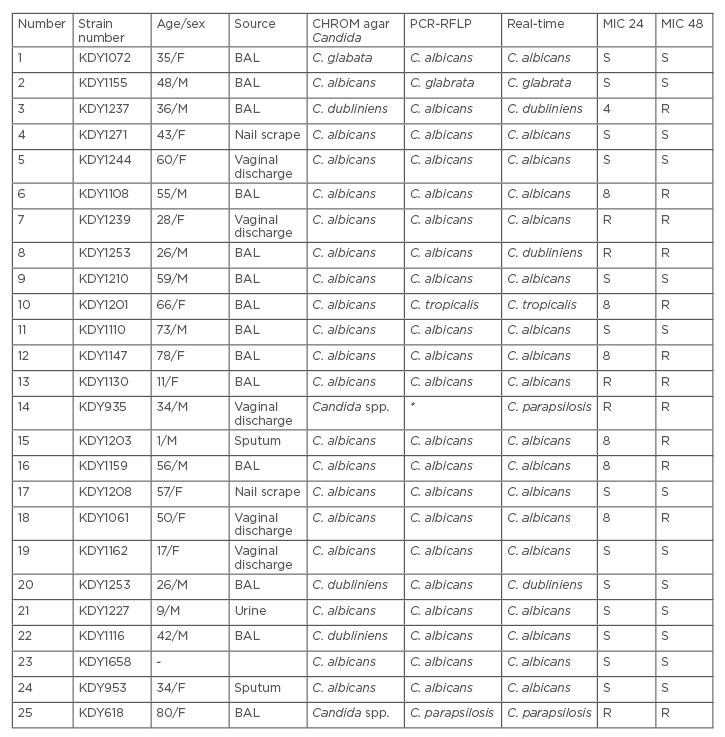
Table 2: Molecular identifications and minimum inhibitory concentration** of itraconazole on clinical Candida isolates.
*Undigested DNA in RFLP profile
**MIC range: 0.063–0.25µg/ml
BAL: bronchoalveolar lavage; F: female; M: male; MIC: minimum inhibitory concentration; R: resistant; RFLP: restriction fragment length polymorphism; S: sensitive.
DISCUSSION
The prevalence of systemic Candida infections, for which hospital care is needed, has recently
elevated significantly for several reasons. Patients at risk of systemic fungal infections (including candidiasis), such as those with a serious underlying disease, immunosuppression caused by cytotoxic/anti-rejection chemotherapy, corticosteroid therapy, and long-term antibiotic therapy, are on the increase.11 Furthermore, fungal infections are recognised as a serious concern amongst the elderly population, as age is a risk factor because of its effect on mortality rates.12,13 Presence of Candida colonisation on the skin surface and mucosa is a potential source of systemic infection, and the hands of medical personnel have been evidenced to play an important role in nosocomial infections with transmission from healthcare workers to patients in hospitals.14 Moreover, transmission from common sources such as contaminated intravenous fluids, hospital food, and medical equipment has been shown.14,15
In the present study, most of the subjects were admitted to specialist medical wards, including nephrology and pulmonology. A significant number of the patients hospitalised were immunosuppressed after a kidney transplant, an important risk factor for Candida colonisation and infection.
Two studies from the USA have shown a medieval prevalence of Candida yeasts at nephrology wards.16,17 This study showed that there was no difference in sex preponderance with regards to either colonisation or infection by Candida yeasts; however, the elderly patients (>50 years old) were more at risk of infection. Similar to other studies, kidney transplant, immune suppression, and haemodialysis were shown to be the main risk factors in this study. Several other studies have indicated that immunosuppression, broad spectrum antibiotic therapy, and diabetes are important predisposing factors of candidiasis.15,17 Another recent study classified antibiotic treatment 3 months after kidney transplant, cytomegalovirus infection, and diabetes as independent risk factors associated with invasive fungal infections.
Amongst all collected specimens in this study, most of the Candida cells were isolated from bronchoalveolar lavage, which indicated Candida colonisation of the mucosal layer of the oropharynx and the lower respiratory tract. Post-transplant immunosuppression may have predisposed the patients to mucosal colonisation by Candida. Several researchers have also reported a high prevalence of Candida species causing pulmonary colonisation infection. This study’s findings of Candida identification showed that C. albicans was the most common yeast isolate. This result is consistent with that of several other studies.17 Another study from the USA reported C. albicans as the most common fungal isolate from 28 kidney transplant, 17 liver transplant, six lung transplant, and four heart transplant patients.9 Nonetheless, non-albicans Candida species, such as C. glabrata and C. dubliniensis, have been identified as leading isolates in other studies. The importance of real-time PCR in this research was highlighted by its ability to differentiate C. dubliniensis, which could not be differentiated by PCR-RFLP and a restriction enzyme. Using real-time PCR for the molecular identification of Candida yeasts has been reported by several researchers. A recent study from China reported the identification of four Candida species, including C. albicans, C.tropicalis, C. glabrata, and C. dubliniensis. Another study from Germany used real-time PCR assays for identifying C. albicans, C. parapsilosis, C. glabrata, C. tropicalis, and C. dubliniensis.10 There were no coincidences amongst the three methods used in the present study, which involved at least seven Candida isolates at the species level. For example, C. tropicalis identified by the CHROMagar™
differential medium was not confirmed by PCR-RFLP and real-time PCR. However, one case of C. albicans identified by real-time PCR was identified as C. glabrata by CHROMagar™ and PCR-RFLP. These results of AFST (MIC) indicated that approximately half of the clinical isolates of C. albicans exhibited itraconazole resistance. Two thirds of cases of C. dubliniensis and both isolates of C. parapsilosis were also resistant. A study from Turkey reported 11/23 isolates of C. albicans and 3/5 isolates of C. glabrata were resistant to itraconazole (MIC: ≥16 μg/mL).12
CONCLUSION
In conclusion, the authors found that immunosuppressions and/or iatrogenic interventions predispose patients hospitalised at medical wards to yeast colonisation/infection. C. albicans is the predominant species amongst the isolates that colonises in the upper respiratory tract. It was also found that C. albicans had a decreasing susceptibility to itraconazole (an azole antifungal drug). Molecular identification of Candida species and their resistance patterns is essential to optimising patient management and reducing unnecessary drug use.