Abstract
The authors present a case report caused by Francisella novicida, a rare opportunistic human pathogen that may cause a tularemia-like disease in patients who are immunocompromised. The diagnosis is a challenge since it can be confused with Pasteurella or Brucella, and matrix-assisted laser desorption ionisation time-of-flight systems are limited due to its poor performance in identification.
Key Points
1. This manuscript describes a clinical case report caused by Francisella novicida, an uncommon bacteria that can cause opportunistic human infections in patients who are immunocompromised, with a range of symptoms from afebrile lymphadenopathy to pneumonia.
2. It is important that microbiologists bear this micro-organism in mind and to alert clinicians, since this disease is rare in the Southern hemisphere and occurs infrequently in patients.
3. The approach to the identification of this species is a challenge since the phenotypic identification cannot be achieved using biochemical tests and is not included in the matrix-assisted laser desorption ionisation time-of-flight mass spectra database.
CASE STUDY
A 38-year-old male with a right cervical mass, fever, and sweating was admitted to the authors’ hospital in 2020. The patient had no weight loss or asthenia.
In 2015, the patient had presented with osteoarticular disseminated histoplasmosis, negative IgM, and negative anti-core antibodies (through a chemiluminescent microparticle immunoassay [CMIA]), which was treated with intravenous (IV) amphotericin B for 7 days, and then with itraconazole (200 mg/12 hour) for 1 year. In 2018, the patient also presented lymph node salmonellosis and was treated with IV ciprofloxacin for 11 days.
On admission a CT scan was performed, showing several enlarged lymph nodes visible on chest, abdominal, and cervical locations. The abdominal CT showed multiple lymph node images in the intercaval left lateral aortic, and coeliac regions, as well as in both external and retrocrural iliac chains.
Laboratory results revealed: haematocrit: 39.8%; white blood cells: 9.490×103 µL; lymphocytes: 8.0% (0.759×103 µL); platelets: 344.000×109 /L; urea: 28 mg/dL; and glucose: 92 mg/dL. The patient had normal transaminases (aspartate transaminase: 29 mg/dL; alanine transaminase: 14 mg/dL), and alkaline phosphatase (356 mg/dL).
In the context of prolonged febrile illness, serology tests were carried out and showed non-reactive protein 24 antigen; anti-HIV-1 and HIV-2 antibodies (CMIA); non-reactive anti-human T-lymphotropic virus 1 and 2 antibodies (ELISA); negative hepatitis B (HB) core antibodies and HB surface antibodies (CMIA); negative anti-hepatitis C antibodies (CMIA); positive IgG anti-cytomegalovirus and negative IgM (CMIA); negative IgM anti-capsid antibodies (enzyme-linked fluorescence assay [ELFA]); negative Epstein–Barr virus; positive anti-capsid IgG antibodies (ELFA); positive anti-Epstein–Barr virus antibodies (ELFA); negative anti-Brucella abortus antibody (Huddleston and Rose Bengal Tests); IgM negative anti Bartonella henselae antibodies; and negative PCR for severe acute respiratory syndrome coronavirus 2.
Because of the patient’s history of infections, their immune status was studied with the following results: cluster of differentiation (CD) 4 cell counts in peripheral blood were 272 cells/mm3 (normal value: 771–1,180 cells/mm3), with a percent value of 30% (reference value: 10–38%); CD8 cell counts in peripheral blood was 229 cells/mm3 (normal value 629–1,128 cells/mm3), with a percent value of 73% (reference value: 55–83%); and CD3 cell counts in the peripheral blood of 550 cells/mm3 (normal value: 1,543–2,484 cells/mm3), with a percent value of 36% (reference value: 28–57%).
A scheduled lymph node biopsy was performed. In the following days, asthenia and temperature increased; therefore, the patient was hospitalised.
The biopsy sample was cultured for aerobic and anaerobic bacteria, as per the procedures of the microbiological laboratory. The conventional method was used in solid media (Löwenstein–Jensen [LJ] and Stonebrink mediums, with and without decontamination) to detect mycobacteria. The sample was also cultured for mycology diagnosis on Sabouraud agar and brain heart infusion blood agar (fungal formulation) at 28 °C and 35 °C.
Cultures for mycobacteria and fungi obtained from the lymph node biopsy were negative at 60 and 30 days of incubation, respectively.
At the bacteriology laboratory, the lymph node biopsy was cultured in chocolate (bioMérieux [Marcy-l’Étoile, France]) and Columbia blood agar. The biopsy was incubated in a 5% carbon dioxide atmosphere at 35 °C, and in Brucella blood agar, with and without antibiotics, in an anaerobic atmosphere. The microscopic examination on Giemsa stain did not show intracellular yeasts and Ziehl–Neelsen stain was negative. Gram stain using safranin was also negative, but the counterstain with fuchsin showed several short Gram-negative rods (Figure 1). Small colonies (1.0–1.5 mm) were observed after 72 hours of incubation on 5% sheep blood agar and on the chocolate agar plates. No differences in colony size were observed between both plates.
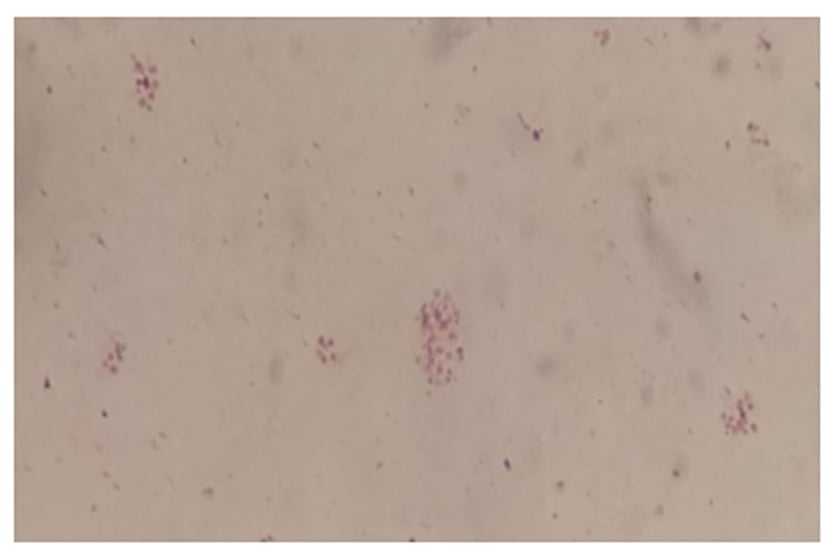
Figure 1: Gram stain of Francisella novicida isolate.
Morphology: very tiny gram-negative pleomorphic coccobacilli.
Since the patient continued to have a fever, three sets of blood culture were obtained, each set including one aerobic and one anaerobic bottle. All sets, including aerobic blood bottles (BACTEC [Becton, Dickinson and Company, Sparks, Maryland, USA]) were positive at 44, 45, and 42 hours. Gram stain using fuchsin showed Gram-negative coccobacilli, identical to the Gram stain observed in the lymph node biopsy culture.
Matrix-assisted laser desorption ionisation time-of-flight (MALDI-TOF Biotyper; Bruker [Billerica, Bremen, Germany]) was also used to identify the micro-organism. Mass spectra were acquired using the MALDI-TOF MS in a linear positive mode (Microflex [Bruker]) in an m/z range of 2,000–20,000, using a Microflex LT controlled by FlexControl (software version: 3.4 [Bruker]).
The isolate could not be identified (not reliable identification) by MALDI-TOF MS. The phenotypic identification was performed by traditional biochemical tests, according to the Wauters and Vaneechoutte scheme.1 The isolate was negative for oxidase, motility, nitrate reduction, urea, and indole; it also showed weak catalase activity. Acid production from carbohydrates glucose, glycerol, and sucrose in a cystine trypticase agar base with phenol red indicator was detected, but not from lactose. Pyrrolidonyl arylamidase and trypsin activity was detected. Gelatine hydrolysis was not detected. No inhibition halo (6 mm) to vancomycin (30 µg) and colistin (10 µg) were observed by disc diffusion on Mueller–Hinton blood agar. These results suggested that the isolate could belong to the Francisella genus (Table 1).1,2
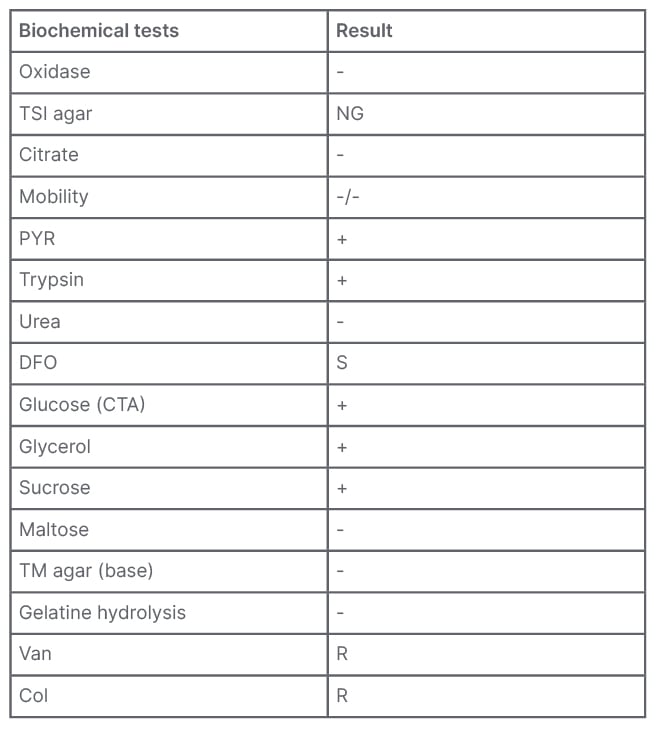
Table 1: Biochemical tests to identify Francisella species.
Col: colistin; CTA: cystine trypticase agar; DFO: deferoxamine; NG: no growth; PYR: pyrrolidonyl arylamidase; R: resistant; S: susceptible; TM: Thayer–Martin; TSI: triple sugar iron; Van: vancomycin; -: negative; +: positive.
Antibiotic susceptibility was determined by epsilometer test (Etest [bioMérieux]) on Mueller–Hinton agar, supplemented with 5% sheep blood agar, and incubated in a 5% carbon dioxide atmosphere at 35 ºC for 24 hours. Minimum inhibitory concentration results were interpreted using the Clinical and Laboratory Standards Institute (CLSI) susceptibility breakpoints for Francisella tularensis with levofloxacin and doxycycline; Pasteurella with ceftriaxone and ampicillin/sulbactam; and Haemophilus influenzae breakpoints with clarithromycin.3
Minimum inhibitory concentration results showed that the isolate was susceptible to all the antibiotics: ampicillin/sulbactam (3.000 µg/mL); ceftriaxone (0.250 µg/mL); clarithromycin (3.000 µg/mL); doxycycline (0.250 µg/mL); and levofloxacin (0.064 µg/mL).
The patient was treated empirically with IV ceftriaxone for 14 days. Doxycycline was initiated 2 days after starting ceftriaxone. At discharge, the patient’s treatment was changed to ciprofloxacin 500 mg every 12 hours, plus doxycycline for up to 3 months.
In order to further identify the isolate, 16S ribosomal (r)RNA and pgm gene were amplified. Total DNA was extracted and used to perform PCR reactions according to the manufacturer’s instructions (Inbio Highway® [Buenos Aires, Argentina]). Specific primers were designed for pgm amplification (pgmF: AGGCTTTTGGTGGGATTGTA; pgmR: AGTTGGTTCAGTCATTCCTGTT). By 16S rRNA gene sequencing, the strain was proven to be F. tularensis. The presence of the F. tularensis subspecies (subsp.) Francisella novicida was confirmed by pgm gene amplification, which showed a 99% identity with F. novicida U112 (AN CP009633). The F. novicida pgm gene sequence was deposited at GenBank (National Center for Biotechnology Information [NCBI; Bethesda, Maryland, USA]) under the accession number OQ122144.
DISCUSSION
The genus Francisella, a member of the γ-subclass of Proteobacteria, contains five valid species isolated in human sources: F. hispaniensis, F. opportunistica, F. salimarina, F. philomiragia, and F. tularensis.4,5 There are currently three proposed subsp., each of which displays several biochemical, epidemiological, and virulence characteristics: F. tularensis subsp. holarctica, F. tularensis subsp. mediasiatica, and F. tularensis subsp. tularensis.6,7 In 2010, Huber et al.8 validated the publication of the name F. tularensis subsp. novicida (herein F. novicida). However, according to the List of Prokaryotic names with Standing in Nomenclature (LPSN),9 F. novicida has not been validly published. It is the correct name if this species is regarded as separate species; however, the appropriate nomenclature for F. novicida has been controversial.
F. tularensis is a Gram-negative coccobacillus and is the causative agent of the zoonotic disease tularemia in humans and animals.7 The subsp. tularensis and holarctica are those most commonly associated with human disease.7 However, F. novicida is considered a rare opportunistic human pathogen,10,11 which may cause a tularemia-like disease in patients who are immunocompromised, similar to F. philomiragia.12
Kingry et al.11 have highlighted clinical, ecological, genomic, virulence, and pathogenic differences between F. novicida and F. tularensis. F. tularensis causes the zoonotic vector-borne disease tularemia, while F. novicida does not. As determined by whole genome comparisons, F. tularensis evolved independently of F. novicida, which is consistent with its completely distinct ecological niche and mechanisms of transmission. Moreover, in relation to their intracellular lifestyle, they have different strategies to evade the immune response. The formation of the inflammasome, a multi-protein complex that is present in the host cell cytoplasm and can be activated by microbial components to induce maturation of cytokines, leading to death of infected cells, is present in F. novicida but not in F. tularensis. Therefore, F. novicida is unable to efficiently evade the host immune response in contrast to F. tularensis.
According to the authors, F. novicida encodes 84 genes that are inactivated in F. tularensis. The predicted function of these genes (carbohydrate metabolism, amino acid biosynthesis, metabolite transport, energy metabolism, transport, and DNA restriction or modification) is consistent with F. novicida maintaining the ability to exist in the environment, outside animal hosts. Genomic analyses of F. tularensis and F. novicida indicate a duplication of the 30 kb Francisella pathogenicity island (16–19 genes comprising a Type VI secretion system) in F. tularensis in comparison to F. novicida, which contains only a single copy. Furthermore, the virulence of F. novicida upon subcutaneous introduction appears to be less than F. tularensis in mice, guinea pigs, and rabbits. In addition, the cell surface, a critical pathogenicity determinant, is different between these two species. The structurally and antigenically unique O-antigens from both species appear to play different roles in the pathogenicity of each strain. Another difference has been observed in pulmonary infection in C57 black 6 mice, which demonstrated dissimilar cell types infected in vivo.11
The diagnosis of F. novicida is challenging. It is difficult to see the cells on Gram stain because they are very small, even smaller than the Pasteurella species, and safranin is not recommended (Figure 1) as a counterstain. On blood and chocolate agars, the colonies are also like Pasteurella, and there is no growth in Levine eosin methylene blue or MacConkey agar since they have nutritional requirements (Figure 2).
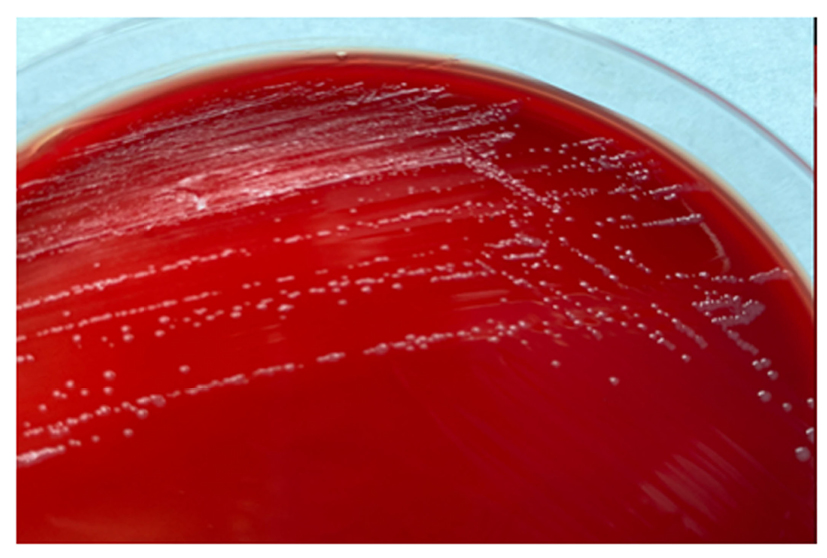
Figure 2: Francisella novicida colonies in blood agar after 72 hours of growth.
Small colonies (1.0–1.5mm) with entire margin, smooth, and moist.
However, the isolate differed from Haemophilus influenzae or Pasteurella since the colonies did not have the typical mouse-like odour that these genera usually have.12 Besides, the isolate did not grow in triple sugar iron agar.
Using standard biochemical tests, the authors ruled out Brucella because the nitrate reduction, urease, and oxidase tests were negative. However, it should be noted that Brucella canis often oxidases negatively, and only the urease test is useful for the differentiation of both.1,12
The MALDI-TOF MS system is limited due to its poor performance in Francisella species identification. The main reason could be attributed to the fact that, in the authors’ laboratory, none of the agents of bioterrorism are included in the database, and F. tularensis is known as a potential biological weapon due to its high virulence and low infective dose. When the authors performed the identification, a reliable identification could not be reached. Several spectra of F. philomiragia have been included in the database, but there is no other species of the genus included in the database. When the authors observed the top 10 identification scores, this species appeared among the options but not with a score that could at least suggest the genus identification.
Even though 16S rRNA gene sequencing is widely accepted as a method for species identification, there are some cases in which the amplification of alternative genes is more suitable for identification.13,14
The fact that F. novicida and F. tularensis share approximately 97% nucleotide identity, could lead to a misidentification between them. Therefore, it has been described that the amplification of different genes (such as pdpD, sdhA, uup, aroA, atpA, pgm, tpiA, trpE, and parC) would be useful for further resolution between F. novicida and F. tularensis.15
In the authors’ case, the sequencing and subsequent analysis of the pgm gene allowed them to correctly identify the isolate.
Human infections caused by F. novicida are rare and considered opportunistic infections. Isolates were recovered from blood, lymph node tissue, and wounds.2 Conversely F. tularensis causes tularemia in healthy individuals, which may be presented with any one of the clinical forms such as: ulceroglandular, glandular, oculo-glandular oropharyngeal, and pneumonic. The port of entry is via an infective arthropod bite (from ticks, flies, or mosquitoes), direct contact with infected animals, ingestion of water or food contaminated by infected animals, and inhalation of infective aerosols. In contrast, F. novicida is not a zoonotic pathogen and, due to its low virulence, infections are unusual. The few cases described occur in patients who are immunocompromised, so its accurate diagnosis is difficult.11 F. novicida has never been identified in arthropod vectors in nature and the only source has been associated with salt water.2,16
Clinical information available of 11 reported cases indicate that nine of the F. novicida cases occurred in patients who were immunocompromised or had underlying health conditions. Clinical symptoms of infection range from afebrile lymphadenopathy to pneumonia.14,17-21 In the two healthy individuals with F. novicida infection, regional lymphadenopathy with no fever or other symptoms was reported.17,19 In these cases, the route of infection was uncertain. Two cases were due to near-drowning events in salt water, and three cases were associated with environmental contamination of outdoor ice machines.15,21
The authors’ patient developed idiopathic lymphocytopenia, multiple enlarged lymph nodes, and fever like glandular tularemia, while HIV was ruled out.
The clinical presentation in the authors’ patient, which included fever and cervical lymphadenopathy, in addition to the above-mentioned opportunistic diseases, led the authors to study the immune system and perform a lymph node biopsy to dismiss a lymphoproliferative process. During the pandemic, the authors’ patient worked in a rural area, helping patients with addictions to recover. Moreover, the authors’ patient did not report any bite, and this was not observed during physical examination. The patient’s medical record indicated they had visited a coastal city 3 months before the onset of symptoms, suggesting an environmental source of the infection.
There is no validated treatment for infections caused by this species, but antibiotics used for tularemia are usually effective for F. novicida infections. Aminoglycosides, tetracyclines, chloramphenicol, and quinolones are frequently used in the treatment and prophylaxis of tularemia.7 Although they rapidly acquire resistance to fluoroquinolones, they have been demonstrated in vitro in both F. tularensis and F. novicida, while natural strains with acquired resistance have not been reported so far.10,22
Despite sensitivity tests being standardised by the CLSI for F. tularensis, for ciprofloxacin, doxycycline, chloramphenicol, and gentamycin the results of the antibiotics tested in vitro were active against this strain. The patient had a favourable progress after antimicrobial treatment.
To the best of the authors’ knowledge, this clinical case is the first report of F. novicida described in Argentina. The approach to the identification of this species is a challenge. It is important that microbiologists bear this micro-organism in mind, since it is rare in the Southern hemisphere and uncommon in patients.








