Abstract
The COVID-19 pandemic has been a huge challenge for the Greek National Health System. Real-time reverse transcription PCR (rtRT-PCR) remains the reference method for early diagnosis, contact tracing, and containment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The aim of this study is the documentation of the epidemiological features of SARS-CoV-2 laboratory surveillance with rtRT-PCR in the population residing in the Pieria province of Greece. Of the 15,486 nasopharyngeal and oropharyngeal samples tested with real-time reverse transcription PCR for the presence of SARS-CoV-2 RNA, 8,051 (52%) were from females and 7,435 (48%) from males, aged 7 days–103 years, with 69.9% coming from the age group of >40 years. The 4,616 out of 15,486 (29.8%) samples came from hospitalised patients. There were 3,771 positive samples out of 15,486 (24.3%); 1,890 (50.8%) males and 1,881 (49.2%) females, with the age group of 40–59 years being dominant (29.9%). Those diagnosed for the first time made up 3,352 out of 3,771 (88.9%) of positive samples. The monthly positivity rate ranged from 6.24–15.69% during the B.1.1.7 variant wave, 17.38–52.89% during the B.1.617.2 variant wave, and 59.76% during the first month of the B.1.1.529 variant wave. Absence of detection of the spike protein gene target was observed in 1,371 (36.4%) of positive samples. Cycle threshold values <20, indicative of higher viral load, had 43.2% of positive samples during the B.1.1.7, 70.0% during the B.1.617, and 92.0% during the first month of the B.1.1.529 wave. The positivity and distribution of variants in the study population was in accordance with the respective results announced by official government authorities for the Pieria region.
Key Points
1. This is the first analytical study that provides useful information regarding the prevalence and spread of different severe acute respiratory syndrome coronavirus 2 variants to the Greek National Health System, thus assisting in better health surveillance.
2. Real-time reverse transcription PCR was used to outline the landscape of positive severe acute respiratory syndrome coronavirus 2 samples, as well as the distribution of different variants in 15,486 people attending local healthcare facilities.
3. Variant identification can assist in the clinical management of patients in healthcare settings and also provide critical information for public health strategies.
INTRODUCTION
COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic by the World Health Organization (WHO) in March 2020.1 The health and diagnostic services of all countries were faced with the urgent need for rapid and reliable detection of the virus from appropriate samples. Mass screening and early diagnosis were, and still are, important factors in identifying and isolating carriers, ideally before symptom onset, contact tracing, limiting the spread of the disease, and effectively confronting the pandemic.2-4
Since the onset of the SARS-CoV-2 pandemic, several variants of the virus have been identified. Depending on their ability to transmit, evade immunity, and disease severity, they have been characterised as variants of concern or variants of interest. Therefore, it is important for public health responses that variants can be identified and specified as accurately as possible regarding their epidemiological characteristics, and impact on the population.5-7
The reference method proposed by the WHO for the diagnosis of SARS-CoV-2 infection is the detection of viral RNA with real-time reverse transcription PCR (rtRT-PCR) in respiratory samples such as nasopharyngeal and oropharyngeal swabs, bronchial aspiration samples, oral swabs (saliva), and sputum.8 Studies have reported the detection of SARS-CoV-2 RNA in blood, serum, and plasma.9,10 However, nasopharyngeal, oropharyngeal, combination (naso-oro-pharyngeal) and oral swabs, and saliva samples, are the most appropriate for molecular diagnosis,11,12 and are recommended by the Centers for Disease Control and Prevention (CDC) and the European Centre for Disease Prevention and Control (ECDC).13,14
From the beginning of the pandemic until the end of 2020, the molecular testing carried out by the Microbiology Laboratory of General Hospital of Katerini (GHK) in Greece was aimed at the diagnosis of emergencies, symptomatic and asymptomatic, and the prevention of inpatient viral dispersion. The largest volume of samples was tested by the microbiology laboratory of the Medical School, Aristotle University of Thessaloniki, Greece. However, during the second pandemic wave, in the autumn of 2020, several factors such as the large number of patients with COVID-19 increased hospitalisation needs in the COVID-19 clinic and intensive care unit, as well as the need for epidemiological surveillance of the regional population for disease dispersion, mandated the development and operation of the Laboratory for the Molecular Diagnosis of COVID-19 at GHK.
In this study, the authors aim to examine and present the demographic characteristics and results of the molecular testing from the laboratory surveillance of SARS-CoV-2 with rtRT-PCR, in order to assess the spread and prevalence of the SARS-CoV-2 variants in Pieria in northern Greece between January 2021–January 2022. However, the study includes only samples from residents who underwent molecular testing at the Molecular Testing Laboratory for Diagnosis of COVID-19 at GHK, and does not include those tested by the mobile units of the National Public Health Organisation (EODY) and private diagnostic laboratories, nor the results of the Ag Rapid tests. To the authors’ knowledge, this is the first analytical documentation of SARS-CoV-2 demographic and laboratory findings concerning a regional population attending the primary public health care facilities in Greece.
MATERIALS AND METHODS
From January 2021–January 2022, the Laboratory for the Molecular Diagnosis of COVID-19 at GHK conducted molecular testing with rtRT-PCR for the presence of SARS-CoV-2 RNA on 15,486 nasopharyngeal and oropharyngeal swab samples from citizens attending regional primary health facilities in Pieria. Demographics, sampling unit, testing reason, and SARS-CoV-2 contact information were obtained from the sample reference document to the degree they were provided. The provided information and rtRT-PCR results were used to assess the spread and prevalence of the SARS-CoV-2 variants in the above population in comparison to national results.
Samples were collected in IMPROVIRAL NAT Medium (Improve Medical Instruments Co. Ltd., Guangzhou, China) or Disposable Virus Sampling Tube (Zybio Inc., Chongqing, China), maintained at 4 ℃ and examined within 24–48 hours of sampling. Samples examined after 48 hours were preserved at -30 ℃ until the day of processing.
Viral RNA was extracted from 200 μL of each sample with the MagMAX™ Viral/Pathogen II (MVP II) Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA), amplified with TaqPath COVID-19 CE-IVDRT-PCR Kit (Thermo Fisher Scientific) in the QuantStudio™ 5 Dx Real-Time PCR System (Thermo Fisher Scientific). Results were obtained after data analysis by the COVID-19 Interpretive Software CE-IVD editions v2.3 and v2.5 (Thermo Fisher Scientific), according to the manufacturer’s directions. All of the above are In Vitro Diagnostic (IVD) certified for in vitro diagnostic use.
The viral load of the samples was estimated by the cycle threshold (Ct) values of the positive samples.
RESULTS
During a 13-month period, 15,486 nasopharyngeal and oropharyngeal samples were tested for the presence of SARS-CoV-2 RNA. Samples were collected from residents of the regional province of Pieria, who attended or were hospitalised at GHK; the psychiatric clinic and its outpatient units; regional primary healthcare units, including the health centres of Aiginio, Litochoro, and Katerini; and the regional clinics for preventive or diagnostic testing. The demographic characteristics of the examinees, the origin of the samples, the reason for examination, and the positivity distribution per group are presented in Table 1, while the monthly percentage positivity distribution per SARS-CoV-2 variant and sampling unit is presented in Table 2. Positivity distribution data per testing reason was not available.
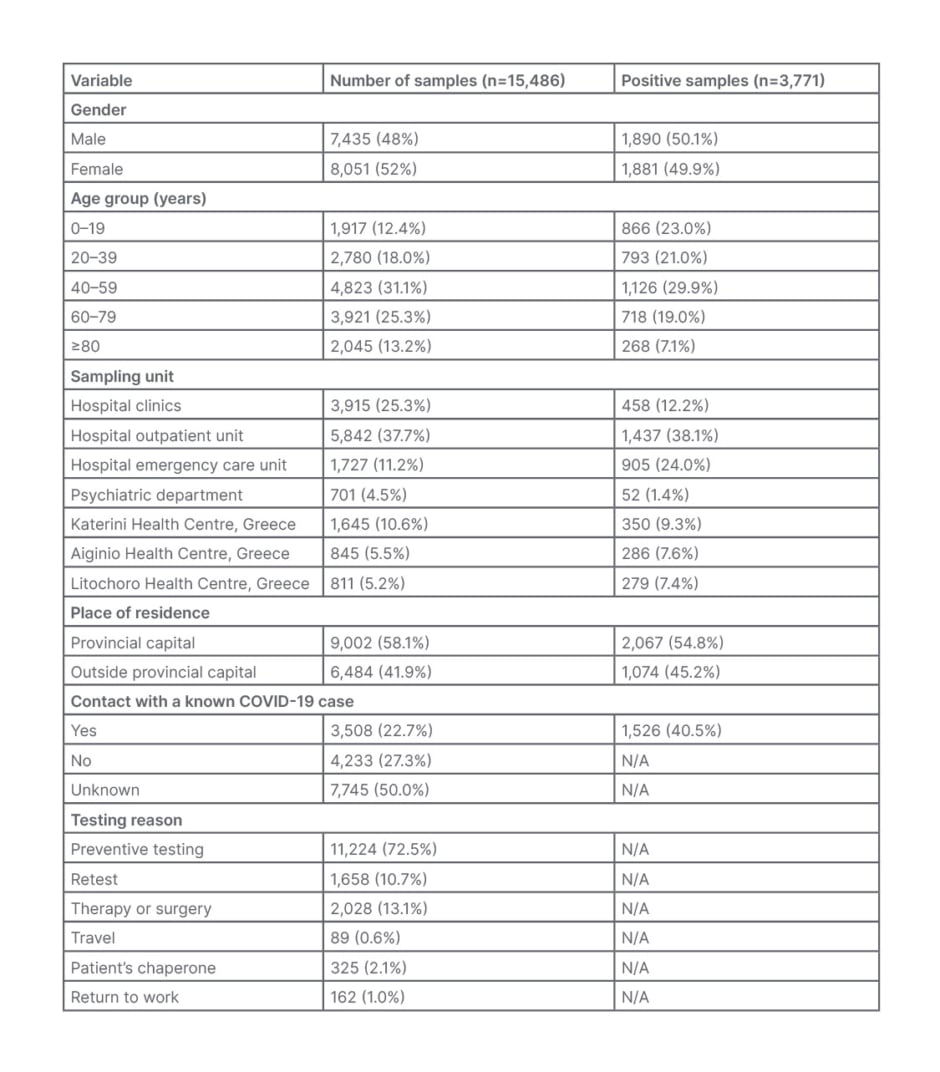
Table 1: Demographic and laboratory characteristics for the 15,486 samples tested at the General Hospital of Katerini, Greece, from January 2021–January 2022.
N/A: no data available.
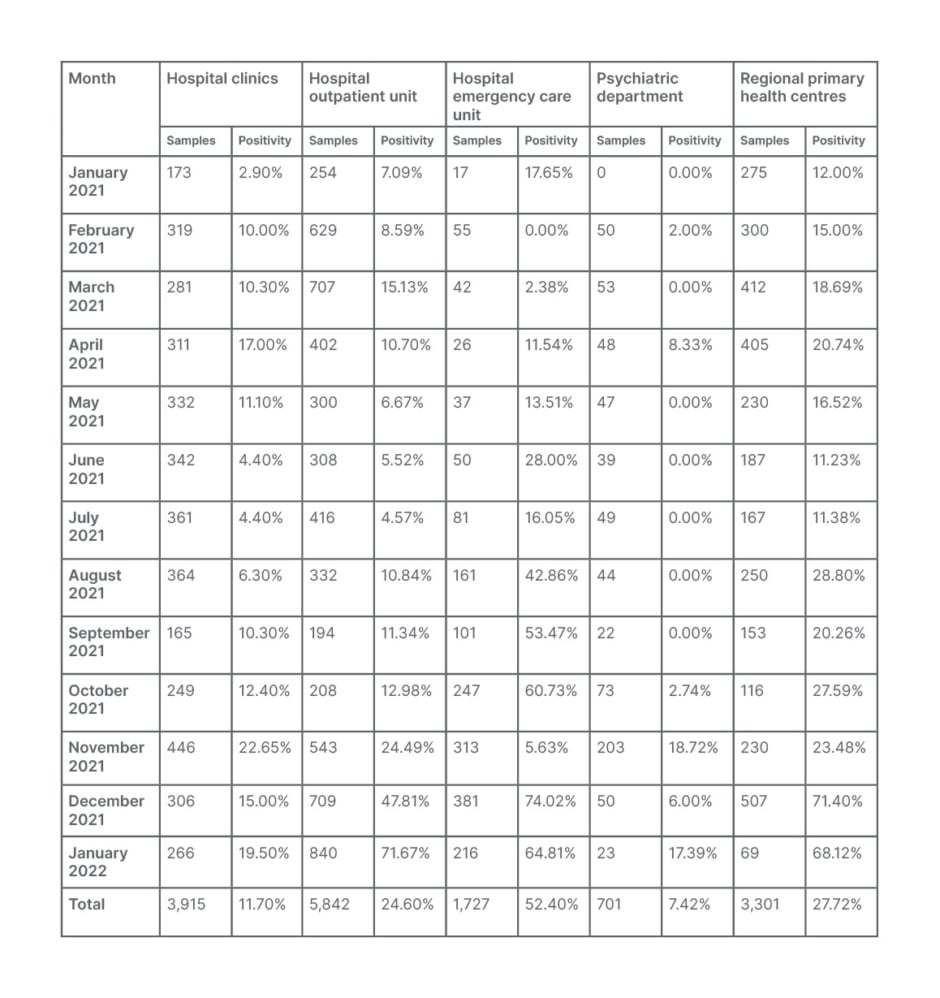
Table 2: Monthly positivity distribution per severe acute respiratory syndrome coronavirus 2 variant and sampling unit for the 15,486 samples tested at General Hospital of Katerini, Greece, from January 2021–January 2022.
The age of the examinees ranged from 7 days–103 years, and those >40 years old contributed 69.9% of the samples. While the majority of the samples were obtained at the hospital settings (12,185 out of 15,486; 78.7%), only 4,616 (29.8%) were taken from hospitalised patients, and the remaining 10,870 (70.2%) from non-hospitalised patients. Regarding contact with a confirmed COVID-19 case, 3,508 (22.7%) of the examinees answered ‘yes’, and 4,233 (27.3%) ‘no’, while 7,745 (50.0%) answered ‘unknown’ or no answer was given. Out of 15,486 samples, 13,828 were examined for the first time, while the remainder concerned follow-up testing of patients and repetitions for the investigation of inconclusive results. Preventive screening for SARS-CoV-2 was the main reason for testing (11,224; 72.5%). Other reasons for testing were re-examination after preventive domestic restriction or illness, screening before therapy or surgery, travelling, return to work, and patient chaperone (Table 1).
Overall, 3,771 (24.3%) samples were positive, with 2,342 (62.1%) of these obtained at the hospital outpatient unit (OU) and emergency care units (ECU; Table 1). Of the 3,771 positive samples, 3,331 (88.3%) were positive for the first time. Of the samples examined, 804 (5.2%) patients had a previous positive molecular result, without specifying the time or the laboratory of diagnosis. Of these 804 samples, 440 (54.7%) concerned hospitalised patients and outpatients who remained positive, while 364 (45.3%) were samples of patients with a negative follow-up result.
The monthly distribution of the sample positivity ranged from 6.2%–59.8% (Figure 1A). During the outbreak of the B.1.1.7 (Alpha) virus variant, positivity ranged from 6.2–15.7%, with maximum positivity (15.7%) in April 2021, and minimum positivity (6.2%) in July 2021. During the B.1.617.2 (Delta) variant wave, the positivity of the samples was much higher (17.4–52.9%), with the highest positivity observed in December 2021. Positivity reached 59.8% at the onset of the B.1.1.529 (Omicron) variant wave in January 2022 (Figure 1A). Regarding the 3,508 patients examined with a confirmed COVID-19 case contact, 1,526 (43.5%) had a positive PCR result. The monthly distribution of the positivity in this group ranged from 19.2% in January 2021 to 48.5% in January 2022.
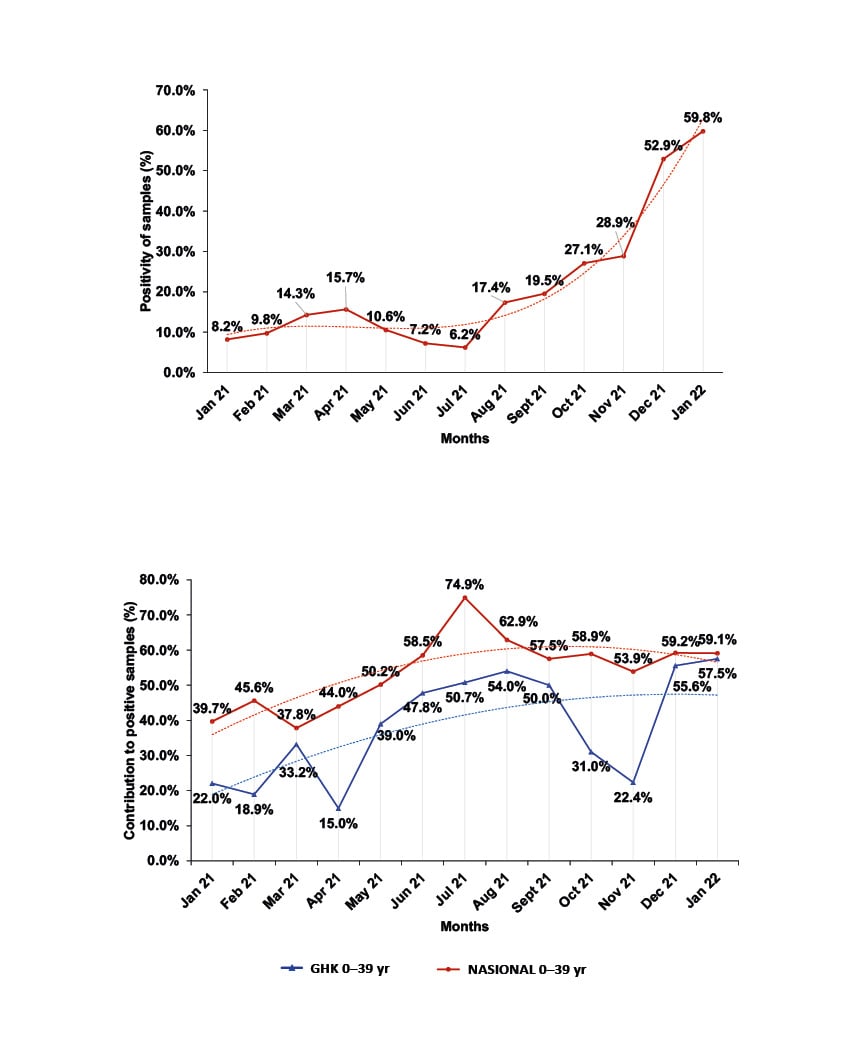
Figure 1: Monthly distribution of sample positivity from January 2021–January 2022.
A) Monthly percentage of the distribution of positivity for the samples tested at GHK.
B) Comparison of the monthly percentage contribution of the age group 0–39 years to positive samples of the province of Pieria, Greece, and at national level.
GHK: General Hospital of Katerini; NASIONAL: national level; yr: years.
Moreover, the monthly positivity distribution per SARS-CoV-2 variant data showed an increase of COVID-19 positive cases in hospitalised patients during the Delta and the onset of Omicron wave (this fluctuated from to 22.65% in November 2021 to 15.00% in December 2021 and 19.50% in January 2022) compared to the Alpha wave (17.00% in April 2021). Similar fluctuations in positivity were observed in patients attending the hospital OU, while the corresponding observation for the hospital ECU, as well as the local primary healthcare centres, show a steady increase of positivity during these periods (Table 2).
The presence of the 69-70del mutation in the spike protein gene (S gene), which prevents the amplification of this target, results in an S dropout effect, also known as the S gene target failure (SGTF). This phenomenon was detected in 1,371 out of 3,771 positive samples (36.4%). Higher SGTF rates were observed in April 2021 (168 out of 187; 89.8%) at the peak of the Alpha variant wave, and in January 2022 (786 out of 845; 93.0%) at the onset of the Omicron variant wave. The lowest (0.0%) were observed in October–November 2021 at the peak of the Delta variant wave (Figure 2).
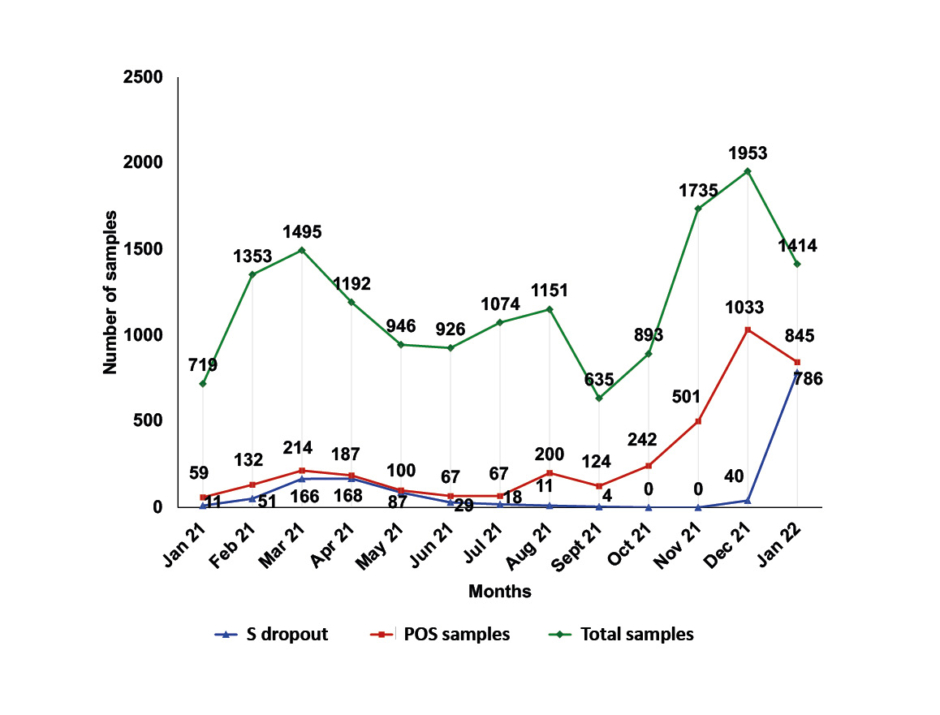
Figure 2: Monthly distribution of samples tested at General Hospital of Katerini, Greece; samples with positive real-time reverse transcription PCR result and samples presenting the S dropout effect from January 2021–January 2022.
POS samples: positive samples.
Ct values of the positive samples in the present study ranged from 9–37. During the Alpha variant wave, 43.2% of positive samples had Ct <20, while during the Delta variant wave the corresponding value was 70.0%, and at the beginning of the Omicron variant wave was 92.0%.
DISCUSSION
There are two limitations in this study, concerning the samples and the provided information to the laboratory. First, the study population does not include those residents tested by the mobile units of the EODY and private diagnostic laboratories in the Pieria province, nor the results of the Ag Rapid tests, but only residents who underwent molecular testing at the Molecular Testing Laboratory for Diagnosis of COVID-19 at the GHK. Secondly, the demographic, contact, and clinical information provided to the laboratory was limited in some cases.
Due to the general data protection regulation, data concerning health facilities of coterminous northern Greece provinces was limited only to the sample positivity obtained from the EODY’s official website.15 As a result, and because of the diversity of rtRT-PCR kits used for the diagnosis of COVID-19 by other healthcare facilities, the authors’ findings could not be analytically compared with data from coterminous health facilities in northern Greece.
The age distribution of the examined citizens, the origin of the samples, and the reason for examination are presented in Table 1. A small predominance of samples obtained from females (8,051 out of 15,486; 52%) was observed over males (7,435 out of 15,486; 48%); this is similar to statistics reported by the EODY for the region of Pieria, provinces adjacent to the Pieria region, and at the national level.15,16 Almost half of the samples examined (7,569 out of 15,486; 48.9%) were obtained at the hospital OU and ECU, and only 4,616 out of 15,486 (29.8%) came from hospital clinics, with the main reason for testing (11,224 out of 15,486, 72.5%) being preventive screening for SARS-CoV-2 (Table 1).
Overall, 3,771 out of 15,486 (24.3%) of the samples were positive, with 3,331 (88.3%) of them obtained from those tested for the first time. Considering that access to hospital services during pandemic waves were restricted to mostly emergency cases, the majority of positive samples (2,342 out of 3,771; 62.1%) came from the hospital OU and ECU (Table 1). The combined positivity for the OU and ECU samples was high (2,343 out of 7,569; 30.9%), and the positivity of the samples obtained at the ECU alone were even higher (905 out of 1,727; 52.4%) as a result of testing mostly citizens with strongly suggestive COVID-19 symptoms. Although 4,616 out of 15,486 (29.8%) of the samples were obtained from hospitalised patients in both clinics and the psychiatric department, including the COVID-19 clinic, only 13.6% of the positive samples came from these patients for two reasons. Firstly, only patients with negative PCR negative results were admitted to non-COVID-19 clinics; secondly, the psychiatric department is comprised of closed and isolated facilities and, therefore, having the lowest positivity rate (Table 1). While in all samples examined there was a slight predominance of females over males, in all positive samples the difference was even smaller (1,890 [50.8%] of males and 1,881 [49.2%] of females), as it was at national level.15,16
Of the samples examined, 804 out of 15,486 (5.2%) had a previous positive molecular result, with 440 out of 804 (54.7%) samples concerning hospitalised patients and outpatients who remained positive, while the remaining 364 out of 804 (45.3%) had a follow-up negative result. The lack of information about the testing time and the testing laboratory for these 364 samples was a limitation to determine the positivity period for these patients. Although the provincial capital population represents the 43.0% of the regional population, it unproportionally contributed 58.1% of the examined and 54.8% of the positive samples (Table 1), probably due to easier access to public and private COVID-19 testing facilities.
While the group of patients <39 years old contributed only to 30.4% of the examined samples, it contributed 44.0% (1,659 out of 3,771) of the positive samples. The positivity rate within this age group was 1,659 out of 4,697 (35.3%), while in the group >40 years, the positivity rate was 2,112 out of 10,789 (19.6%). This difference could be explained as a result of the higher mobility of younger people, the beginning of in-person education after September 2021, and delayed admission to vaccination programmes.15,16 Of those with contact to a confirmed COVID-19 case, 1,526 out of 3,508 (43.5%) had a positive PCR result. The monthly distribution of the positivity in this group of people ranged from 19.2% in January 2021 to 48.5% in January 2022. It gradually increased from 19.2% in January 2021 to 36.1% in August 2021, reaching 47.4% in December 2021, and peaking at 48.5% in January 2022; this is indicative of the ease of viral transmission and increased spread of Delta and Omicron variants.
Another corroborating finding is the overall monthly percentage positivity distribution per SARS-CoV-2 variant and sampling unit, as presented in Table 2. As shown, a wider spread of the disease was observed during the Delta and the onset of Omicron waves compared to the Alpha wave. Moreover, the increase in the number of ECU samples tested, and the corresponding increase in their positivity should be noted, indicative of the increasing number of symptomatic patients attending the hospital ECU during this period. On the contrary, the number of the clinical samples tested and their positivity in January 2022, at the onset of the Omicron wave, were much lower than the respective ones from the OU and ECU, compared to those in April 2021 (Alpha wave) and November–December 2021 (Delta wave; Table 2).These observations are indicative of the increased transmissibility but milder symptoms, resulting in the decreased need for hospitalisation during the Omicron wave compared to the Alpha and Delta variant waves.5,7
The overall monthly distribution of the sample positivity ranged from 6.2–59.8% (Figure 1A). Despite any fluctuations, it showed a sharp increasing trend during the present study, and was much higher than the official reported by EODY for the region.12,13 During the outbreak of the B.1.1.7 variant of the virus, positivity ranged from 6.2–15.7%, with maximum positivity (15.7%) in April 2021 and minimum (6.2%) in July 2021. During the pandemic wave of the B.1.617.2 variant, the positivity of the samples was much higher (17.4–52.9%), with the highest positivity observed in December 2021 at the peak of this pandemic wave. Positivity reached 59.8% on the onset of the pandemic wave of the B.1.1.529 variant in January 2022 (Figure 1A), confirming the higher infectivity and transmissibility of these two variants.5-7 These values, although much higher than those officially announced by EODY for the region, show a similar increasing trend.15,16
Positive samples in the age group 40–59 years were the highest of all (1,126 out of 3,771; 29.9%), comprising 27.7% of males testing positive and 32.0% of females testing positive (Table 1). Positive samples in the age group <19 years were higher compared to the respective reported by EODY at the national level in March 2021 (14.5% versus 8.0%); July 2021 (29.9% versus 16.3%); December 2021 (26.7% versus 19.2%); and January 2022 (35.3% versus 24.6%). In contrast in this age group, during November 2021, while the pandemic wave of Delta variant was evolving, the participation rate was quite low (10.0%) compared to the previous periods, and to the respective at the national level (23.6%) for the same month. Nevertheless, this increase is in concordance with the increase of positivity both at regional and national level during these months.15,16
The above deviations of the positivity in the population and of the participation rates of the age groups in the positive samples are probably the result of two limitations. Firstly, the origin of the samples in the present study includes only the results of molecular testing carried out at GHK and does not represent the general population of Pieria; nor does it include the Ag Rapid test results which are included in the official data of EODY, and which were vastly increased over the PCR tests from April 2021 throughout this study.15,16 Secondly, it does not take into consideration the possible differences in positivity due to quarantine restrictions and the different vaccination timetable for age subgroups. However, it should be noted that throughout the study period there has been an increasing trend in the participation of the age group <39 years in the positive samples commensurate to its participation at the national level and, therefore, a corresponding decrease in the participation of the >40 years age group,15 even though the corresponding percentages in the regional population are lower (Figure 1B). This, in fact, indicates the spread of Delta and Omicron variants among the age group <39 years due to their increased transmissibility, but also due to the initiation of in-person teaching at all levels of education, the relaxation of restrictive measures, and the low vaccination coverage of young people.5-7,15
With the emergence of the B.1.1.7 variant, the mutation 69-70del S was observed in the S gene, which prevented the PCR amplification of the S target. The same mutation was subsequently identified in the B.1.1.529 variant. The SGTF phenomenon, however, does not prevent the interpretation of the results, nor does it make a result negative. On the contrary, this pattern helped in the early recognition of Alpha and Omicron variants, as highlighted by the ECDC and CDC.12,13,16 This pattern was detected in 1,371 out of 3,771 positive samples (36.4%). Higher SGTF rates, as expected, were observed in April 2021 (168 out of 187; 89.8%) at the peak of the Alpha variant wave, and in January 2022 (786 out of 845; 93.0%) at the onset of the Omicron variant wave. The lowest (0.0%) were observed in October and November 2021 at the peak of the pandemic wave of the Delta variant (Figure 2).17
The interpretation of the COVID-19 test results and the evaluation of the SARS-CoV-2 viral load of positive samples were carried out using the Ct value, a semi-quantitative parameter of rtRT-PCR. The Ct represents the lowest number of PCR amplification cycles required to produce a fluorescent signal greater than the background noise. It can approximately indicate the amount of the viral RNA present in the sample; the lower the Ct the higher the concentration, while a higher Ct indicates a low concentration of viral RNA. The Ct values of the positive samples in the present study ranged from 9–37. During the Alpha variant wave, 43.2% of positive samples had Ct <20, while during the Delta variant wave the corresponding value was 70.0%, and at the beginning of the Omicron variant wave it was 92.0%. This is an indication of higher viral loads in the samples of citizens infected with the Delta and Omicron variants, and reflects the higher infectivity of these variants.6,18
However, care should be taken when using Ct values as an indicator for viral RNA load for several reasons. Respiratory tract samples are not homogeneous, like those of blood, urine, or other body fluids. Thus, a nasopharyngeal or oropharyngeal sample with low Ct is usually associated with high infectivity or acute phase infection. On the contrary, a sample with high Ct could refer to a lower-risk, an early, or a rebound infection, but it also could refer to poor sampling procedure (e.g., insufficient collection and storage, or degradation of sample).19-26 In addition, diagnostic laboratories use different reagent kits for RNA extraction and rtRT-PCR, and, therefore, Ct values are not comparable.20,23 It is also known that in some patients a positive rtRT-PCR result is obtained for a long period after clinical symptoms subside (e.g., 60 to 100 days), indicating the shedding of SARS-CoV-2 RNA without necessarily the presence of infectious virus.27-29
Thus, it is obvious that, at present, no consensus exists concerning which Ct values relate to the level of severity of the disease, or to whether or not a person is infectious, unless repeated measures of Ct are taken for each patient throughout the course of the disease.9,30 Therefore, appropriate caution should be taken to interpret the Ct values, and other laboratory and clinical findings should be considered in order to determine a true positive (viable viral shedding) or a true negative person with COVID-19, and the course of the disease.
CONCLUSIONS
In this study, the authors attempted to investigate and present the demographic characteristics and the laboratory molecular testing results of nasopharyngeal and oropharyngeal samples obtained from the residents of the Pieria region, who attended the regional public health care facilities for SARS-CoV-2 testing, and were examined by the COVID-19 Laboratory for the Molecular Diagnosis of COVID-19 at GHK from January 2021–January 2022 inclusive.
In this study, no difference in the positivity of males and females was observed. Sample positivity was lower (6.24–15.69%) during the pandemic wave of the Alpha variant, compared to that of Delta (17.38–52.89%) and the beginning of the Omicron waves (59.76%); this is in accordance with the transmissibility and infectivity of the variants, and the viral load of the samples. Positivity was higher compared to the daily COVID-19 reports of EODY for the Pieria region, as the study only includes the results of molecular testing carried out at the GHK, and does not include results from molecular testing carried out by the testing units of EODY and private laboratories, or the Ag Rapid test results.
In the age group <39 years, there is an increasing trend in positivity from January 2021–January 2022, similar to that at the national level, even though is lower than that. This is indicative of the higher infectivity and transmissibility of Delta and Omicron variants combined with the initiation of in-person teaching at all educational levels, the relaxation of restrictive measures, the high mobility of this age group, and the low vaccination coverage of younger people.
Although the Ct value is a semi-quantitative parameter of rtRT-PCR, the lower Ct values of positive samples with the Omicron variant, compared to Alpha and Delta, are a strong indication of the high viral load, and confirm the increased infectivity and transmissibility of this strain.
The large number of samples tested, including confirmed, probable, and suspected cases of COVID-19, and the ability to distinguish viral variants due to the S dropout effect, could provide the clinicians with important information for the clinical management and hospitalisation of patients. In addition, the detailed documentation of demographic and contact data of the population at the sampling points, and the variant distinction by rtRT-PCR, would be beneficial in providing a comprehensive picture of the prevalence of the disease to public health policy officials, thus contributing effectively to establishing preventive and treatment strategies, to monitoring transmission clusters, contact tracing, and containment of COVID-19 in the Pieria region. Furthermore, the authors’ findings in correlation with those from the primary health care facilities in neighbouring provinces of northern Greece could also provide accurate information of SARS-CoV-2 variant spread in the region during the pandemic phases.








