BACKGROUND AND AIMS
Cholangiocarcinoma (CCA) is a clinically1,2 and molecularly heterogeneous disease.3,4 The ability of microRNAs (miRs) to target multiple signalling pathways links them to tumour heterogeneity.5-7 Here, the authors analysed the miR landscape of CCA and non-malignant livers to define their roles in orchestrating gene regulation and promoting CCA development.
METHOD
The authors performed miR sequencing (miRseq) of 190 human samples (99 intrahepatic CCAs, 28 extrahepatic CCAs, and 63 matched adjacent livers), including matched transcriptome data from 184 samples, to study miRs and regulation of target genes in CCA. High-throughput screening (HTS) using a library of 2,700 miR mimics was performed to characterise their proliferative impact in three primary normal human cholangiocyte cultures. After miRseq-HTS data integration, the authors performed target prediction and pathway over-representation analyses. The main target was confirmed by luciferase reporter assay and characterised by quantitative PCR, immunoblotting, and in situ hybridisation in tissue microarrays. To identify the biological role, the authors generated miR-CRISPR knock-out lines and performed proliferation and wound-healing assays.
RESULTS
MiRseq revealed 398 differentially expressed miRs (388 upregulated, 10 downregulated) in CCA compared with non-malignant tissues. Unsupervised hierarchical clustering identified three tumour subgroups, differing significantly based on tumour location, overall survival, tumour microenvironment (i.e., macrophage, hepatic stellate, and endothelial cell content), and immune infiltrates (dendritic cells, CD4+, and CD8+ T cells).
In HTS, overexpression of 224 miRs increased the normal human cholangiocyte proliferation rate, of which 35 miRs were upregulated in CCA. Pathway over-representation analysis showed FoxO signaling as the major affected pathway.
Further, miR-27a-3p and FoxO1 interaction revealed a strong negative correlation in the CCA cohort, which was independently validated in an external cohort (TCGA-CHOL). This interaction was confirmed in vitro by luciferase reporter assay. In situ hybridisation showed that miR-27a-3p is highly expressed in tumour cells and vascular smooth muscle. MiR-27a-3p knock-out CCA cells showed decreased proliferation and migration, highlighting miR-27a-3p as an oncogenic dependency in CCA.
CONCLUSION
MiRs are highly deregulated in CCA. MiR-27a-3p targets FoxO1, contributing to altered FoxO signalling and tumour phenotypes (Figure 1).
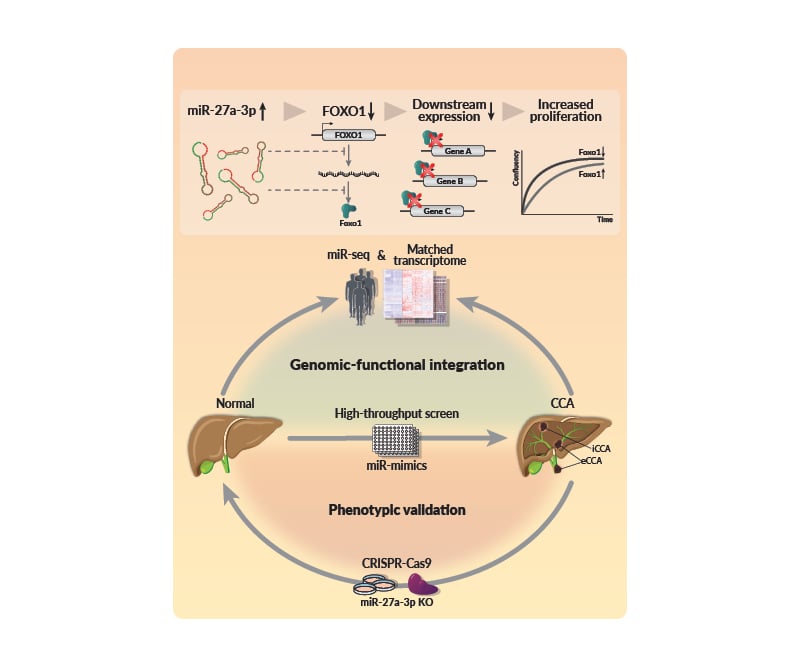
Figure 1: miR-27a-3p regulation of FOXO1 in cholangiocarcinoma.
CCA: cholangiocarcinoma; eCCA: extrahepatic cholangiocarcinoma; iCCA: intrahepatic cholangiocarcinoma; KO: knock out; miR-seq: microRNA sequencing.