BACKGROUND AND AIMS
Despite the fact that activating mutations in RAS-ERK cascade genes are quite often detected in multiple myeloma (MM), the literature data on their prognostic value are contradictory.1,2 The tumour substrate should not only be analysed in the bone marrow and plasmacytoma, but also in the plasma circulating tumour DNA (ctDNA) for the heterogeneity of MM to be effectively analysed.3,4 The aim was to study the mutational status of KRAS, NRAS, and BRAF genes in the tumour substrate from different loci in MM.
MATERIALS AND METHODS
The single-centre study from October 2021–January 2023 included 70 patients with symptomatic MM (29 male, 41 female) aged 35–84 years (median: 58 years). Plasmocytomas were detected in 66% of the patients with MM according to CT data. They were detected in the bone of 40 patients and extramedullary in six. A fluorescence in situ hybridization (FISH) study of CD138+ cells was performed using DNA probes to detect translocations of 14q32/IgH, 8q24/MYC; deletions of 17p13/TP53, 13q14, 1p32; amplification of 1q21; and multiple trisomies (MetaSystems, Altlussheim, Germany). Upon detection of t(4;14) translocation, t(14;16) translocation, del17p13, and amplification of 1q21, the patient was assigned to a high cytogenetic risk group. DNA was isolated from samples of various localisation: CD138+ bone marrow cells (n=60), ctDNA (n=19), bone plasmacytoma (n=9), and extramedullary plasmacytoma (n=6). The mutational status of KRAS, NRAS, and BRAF genes was studied in the tumour substrate from different loci. KRAS and NRAS gene mutations were identified by Sanger sequencing on the Nanophor 05 genetic analyser (Institute for Analytical Instrumentation Russian Academy of Science, Saint Petersburg, Russia), and by next-generation sequencing on the MiSeq System genetic analyser (Illumina, San Diego, California, USA). The BRAF V600E mutation was determined by real-time allele-specific PCR with the device CFX96 Touch (Bio-Rad Laboratories Inc., Hercules, California, USA).
RESULTS
KRAS gene mutations were detected in 16% of patients (11/70), of which less than one-third (27%) had high-risk cytogenetic abnormalities. NRAS gene mutations were detected in another 16% of patients, while more than half (55%) were assigned to a high cytogenetic risk group. BRAF gene mutations were found in 9% of patients (6/70), one-third of whom had high-risk aberrations (Figure 1). Paired tumour samples (plasma ctDNA and CD138+ bone marrow cells) were analysed in 15 patients with MM. In 11 patients, mutations in any of the three genes were found in the bone marrow, while in five patients (45%) similar mutations were also detected in a paired sample of tumour ctDNA isolated from plasma. No cases with KRAS, NRAS, or BRAF gene mutation detected in the plasma and the absence of the corresponding mutation in the bone marrow were found. The mutational status of the three genes was analysed in 15 plasmacytoma samples (nine bone, six extramedullary). It turned out that only KRAS gene mutations (7% of cases) were detected in the samples of bone plasmacytomas, and only NRAS gene mutations (50% of cases) were detected in the samples of extramedullary plasmacytomas.
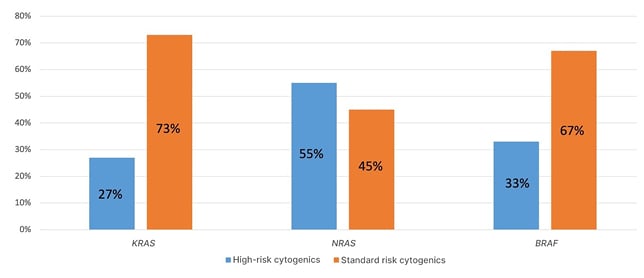
Figure 1: Cytogenetic abnormalities in patients with multiple myeloma with KRAS, NRAS, or BRAF gene mutations.
CONCLUSION
There was a trend towards higher frequency of high-risk cytogenetic aberrations in patients with NRAS gene mutations compared to patients with KRAS gene mutations (55% versus 27%). It was also determined that the NRAS gene was mutated in 50% of extramedullary plasmacytomas samples. In 45% of the cases with KRAS, NRAS, or BRAF gene mutation detected in the bone marrow substrate, similar mutations were also detected in the tumour ctDNA isolated from plasma.






