Despite the high efficacy and improved clinical response of nilotinib in chronic myeloid leukaemia (CML), there is still a significant proportion of patients who fail to obtain or maintain a major molecular response (MR), defined as ≥3 log reduction from the standardised baseline.1 For these patients, treatment-free remission once in deep molecular response (DMR), which is one of the goals of nilotinib therapy, will never be an achievable endpoint. Although BCR/ABL mutations are the major contributory factor for the loss of response to tyrosine kinase inhibitors (TKI), the reduced bioavailability of TKI in leukaemic stem cells is also an important pharmacokinetic factor.2 In this regard, the presence of polymorphisms in drug transports may contribute to mechanisms of drug resistance, disease progression, or loss of MR.
Single nucleotide polymorphisms (SNP) affecting gene expression or function, in both normal and cancerous cells, can cause inherent inter-individual differences in the metabolism and disposition of TKI. Genetic polymorphisms in ABC transporter genes are likely to influence intracellular drug delivery, and, therefore, the effectiveness of TKI.3 It has been documented that ABC multidrug transporter (MDR-ABC proteins) overexpression contributes to imatinib and novel agent asciminib resistance.4 However, less is known about how genetic variants in ABC genes may modify pharmacological properties and affect the response to second- generation TKI.
This study examined five SNP in three ABC transporter genes: ABCC1 5463T>A, ABCC2 3972C>T, ABCC2 rs4148386, ABCC2 1549G>A, and ABCB1 3435C>T, to determine the achievement and loss of molecular responses in CML patients treated with nilotinib. A total of 71 CML patients (40 male and 31 female) with a median age of 50 years were enrolled in the study (Table 1). The genotypes were analysed by PCR-HRM (high resolution melting) assay and PCR-pyrosequencing assay, as previously reported in Visani et al.5 It was noted whether the polymorphisms showed any deviations from the Hardy–Weinberg equilibrium. Differences in genotype and allele distributions among the CML patients and the associations between genotypes with good response, resistance, or loss of response to nilotinib was assessed by Fisher’s exact test, the Kaplan–Meier method, and log-rank test (SPSS, IBM, Chicago, USA; SNPStats package, Bioconductor, Buffalo, New York, USA).
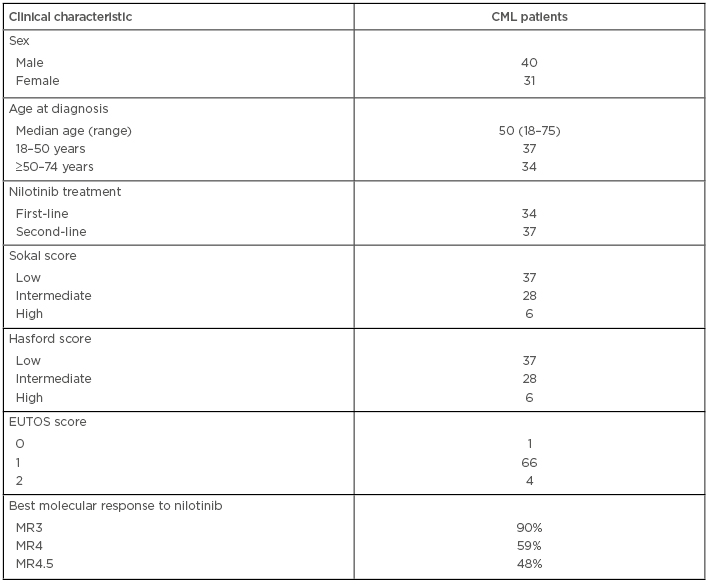
Table 1: Characteristics of patients.
CML: chronic myeloid leukaemia; MR: molecular response.
It was found that the ABCC2 3972C>T (rs3740066) SNP significantly impacted the loss of MR3 in dominant, codominant, and recessive models. Moreover, different genotypes and allele frequencies of rs3740066 in ABCC2 were found in patients who maintain the MR3 and in those who lose it. Patients with the T/T genotype in ABCC2 gene lost the MR3 more frequently than patients with C/C or C/T genotypes in dominant and codominant model (p=0.02; odds ratio [OR]: 11.56 (95% confidence interval: 1.70–78.46); p=0.01, OR: 5.61 (95% confidence interval 1.36–23.09), respectively).
In conclusion, the findings of this study show that the ABCC2 (rs3740066) SNP is associated with a higher incidence of MR3 loss in CML patients treated with nilotinib. A number of previous studies have suggested that individual genetic differences in ABC transporters play an important role in the efficacy and adverse effects produced by drugs. In particular, it has been hypothesised that genetic variants in ABC transporters may alter the pharmacokinetics of TKI and, consequently, may modify the therapeutic response.6,7 Now we are analysing a larger series of CML patients treated with nilotinib and we have extended the genotype analyses to also include SNP in ABCG2, to try to find a possible association between genetic variants and molecular response in CML patients treated with nilotinib.







