BACKGROUND AND AIMS
The expression and tight regulation of microRNA is essential for haematopoiesis and the prevention of leukaemia.1,2 MIR139, which is located in intron-1 of the PDE2A host gene, is a critical tumour suppressor in human cancers and is commonly inactivated in acute myeloid leukemia (AML).3,4 However, the mechanism of MIR139 silencing is still elusive. This study aimed to investigate the effects of MIR139 reactivation on MLL-AF9 AML, to identify the downstream miR-139 targets, and to unravel the molecular mechanism behind the silencing of MIR139 in MLL-AF9 AML (Figure 1).
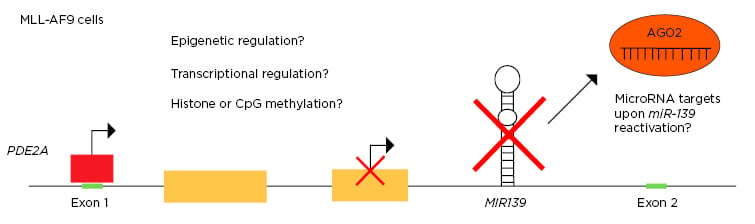
Figure 1: Schematic overview of research questions.
This project aimed to investigate the MIR139 silencing mechanism in MLL-AF9 AML and to identify the miR-139 targets that are controlled upon MIR139 reactivation.
MATERIALS AND METHODS
For this study, the authors selected the AML model with translocation t(9;11)(p22;q23), expressing the mixed-lineage leukaemia (MLL)-AF9 oncogene, which is an aggressive type of AML characterised by poor prognosis.5 miR-139-inducible lentiviral vectors, next generation sequencing, and proteomics were utilised to determine the tumour-suppressing functions and to identify downstream targets of miR-139 in MLL-AF9 AML. To investigate the molecular MIR139 silencing mechanism, the authors employed clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 sgRNA screens, small-molecule inhibitors, CRISPR-Cas9 knockout mouse models, and performed chromatin immunoprecipitation sequencing (ChIP-seq) data mining, RNA interference, and ChIP-polymerase chain reaction.
RESULTS
The study found that MLL-AF9 expression in haematopoietic stem and progenitor cells caused an 85% reduction of miR-139 expression. Forced expression of miR-139 with doxycycline-inducible expression vectors in MLL-AF9 AML caused loss of colony-forming capacity and rapid apoptosis. Next generation sequencing and proteomics revealed Eif4g2, a known miR-139 target, and three novel targets, Ptprt, Map2k6, and Hpgd of miR-139 in MLL-AF9 AML. The study found that CRISPR-Cas9-mediated knock out of Ptprt and Hpgd reduced the colony-forming capacity of MLL-AF9 AML, indicating that these targets mediate the tumour-suppressing activity of miR-139.
Only Eif4g2, Ptprt, and Hpgd were confirmed as direct targets of miR-139 by luciferase-based assays in HEK-293T cells, suggesting that the regulation of Map2k6 is indirectly affected by miR-139. The study found that two well-conserved putative enhancer regions, located upstream of MIR139, are epigenetically silenced by the polycomb repressive complex 2 (PRC2), demonstrated by SUZ12 occupation, increased H3K27me3, and decreased H3K27Ac levels. PRC2 prefers binding to methylated CpG.6 Indeed, the study found increased CpG methylation levels in the enhancer regions, compared to the upstream putative promoter. Inhibiting H3K27-specific methyltransferases enhancer of zeste-1 (EZH1) and EZH2, two critical compounds of PRC2, with UNC19997 resulted in a large increase of miR-139 expression and apoptosis of MLL-AF9 AML, but did not change the expression of Pde2a. Furthermore, results showed that both individual enhancers were essential for miR-139 induction, but not for Pde2a expression in MLL-AF9 cells treated with UNC1999. In agreement, CRISPR-Cas9-mediated deletion of the individual MIR139 enhancer regions in mice resulted in strongly reduced miR-139 levels in normal haematopoietic stem and progenitor cells. Genome-wide CRISPR-Cas9 sgRNA library screens revealed the transcriptional pausing factor of RNA polymerase II, POLR2M,8,9 as a novel MIR139-silencing factor downstream of PRC2. In agreement, Polr2m deletion in MLL-AF9 AML induced miR-139 expression and decreased the colony-forming capacity and cell viability. Furthermore, ChIP experiments revealed that POLR2M is located at the transcriptional start site within the most-proximal enhancer region of MIR139. This interaction, accompanied by enhanced serine 5 phosphorylation of the C-terminal domain of RNA polymerase II, is indicative of paused transcription.10
CONCLUSION
Together, this study indicates that both miR-139 expression, or inactivation of miR-139 targets, eliminate MLL-AF9 AML. Furthermore, the study presents evidence for a POLR2M-mediated MIR139-silencing mechanism downstream of MLL-AF9 and PRC2.







