BACKGROUND
Chronic myeloid leukaemia (CML) is a clonal myeloproliferative disorder with heterogeneous biological and clinical features. Specifically, it presents a reciprocal association between chromosomes 9 and 22, yielding the BCR-ABL fusion protein with overacting tyrosine kinase activity.1 CML is treated with tyrosine kinase inhibitors (TKI), which have dramatically improved the long-term survival of CML patients to approximately 80%.2 Among TKI, nilotinib is very effective for the treatment of imatinib-resistant patients in the clinic.3 However, despite the remarkable success of TKI in controlling CML in the chronic phase, some patients develop drug resistance. The incapability of TKI to eradicate the disease completely is best explained by intrinsic and acquired drug resistance in leukaemic stem cells.4 The main research focus in the CML field is therefore to identify pathways and genes that contribute to the survival of leukaemic stem cells.
AIMS
We performed gene expression profiling (GEP) of selected bone marrow (BM) CD34+/Lin- cells of patients with chronic phase (CP)-CML at diagnosis versus 12 months of nilotinib treatment to gain new insights into the molecular mechanisms of nilotinib treatment in CML.
METHODS
We selected and counted BM CD34+/Lin- cells of 30 CP-CML patients at diagnosis and during 3, 6, and 12 months of first-line nilotinib treatment. BM CD34+/Lin- cells were isolated by immunomagnetic separation technology and tested by standard fluorescence in situ hybridisation (FISH) for all 30 patients at diagnosis and after 12 months of nilotinib treatment. GEP analyses of selected BM CD34+/ Lin- cells of the CP-CML patients at diagnosis and after 12 months of nilotinib treatment were performed by AffymetrixTM (Thermo Fisher Scientific Ltd., Waltham, Massachusetts, USA) microarray analysis. Data were preprocessed using the ComBat method to adjust for batch effects and quantile normalisation.5 The significance analysis of microarrays (SAM) test was used for expression analysis at diagnosis versus 12 months of treatment.6 Selection was performed using the R statistical computing software. False discovery rate-adjusted p values <5% were considered significant.7
RESULTS AND CONCLUSION
We demonstrated that the number of BM CD34+/Lin- cells dramatically decreased after 3, 6, and 12 months of nilotinib treatment (Figure 1). FISH analysis detected CD34+/Lin- Ph+ cells in 30 CP-CML patients at diagnosis, while no Ph+ nuclei were detected in CD34+/Lin- cells after 12 months of nilotinib. GEP detected 264 statistically significant differentially expressed genes at diagnosis versus 12 months of nilotinib treatment. Functional enrichment analysis showed that groups of genes belonging to 14 pathways were significantly dysregulated in CP-CML patients after 12 months of nilotinib. Lipid, glucose, and sphingolipid metabolism, insulin resistance, complement and coagulation, platelet activation, the cytoskeleton, cell adhesion, cellular transport, B cell differentiation, the RAS signalling pathway, proliferation, growth factors, and apoptosis were all shown to be significantly altered after 12 months of nilotinib in comparison with diagnosis in CP-CML patients.8,9
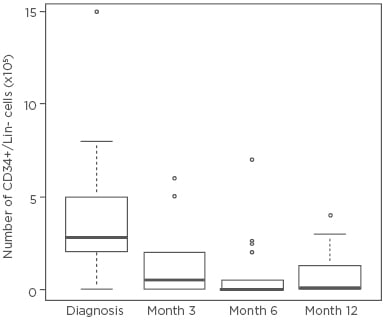
Figure 1: The number of bone marrow CD34+/Lin- cells at diagnosis and after 3, 6, and 12 months of nilotinib treatment in 30 chronic myeloid leukaemia patients.







