Meeting Summary
The 2022 International Scientific Conference of Probiotics, Prebiotics, Gut Microbiota and Health® took place in Bratislava, Slovakia, from 27th–30th June. A common theme throughout the conference was that, although significant progress has been made, considerable work remains to be done in understanding how probiotics work, aligning definitions of microbiome-modifying agents, and communicating evidence-based recommendations to clinicians and consumers. There was also an overarching concern around the ease with which food supplements receive marketing approval compared to drugs, and where probiotics lie on this spectrum.
Arthur Ouwehand, International Flavors & Fragrances, Inc. (IFF), Kantvik, Finland, presented updates to the 2022 International Life Sciences Institute (ILSI) Concise Monograph on probiotics, prebiotics, and the gut microbiota in human health. Hania Szajewska, Medical University of Warsaw, Poland, focused on clinical recommendations for the use of probiotics, suggesting ways to approach the differences between guidelines. Regulatory agencies now recognise probiotics as a new category of medicinal products termed ‘live biotherapeutic products’, and Sin-Hyeog Im, Pohang University of Sciences and Biotechnology, Pohang, South Korea, and ImmunoBiome Inc., Pohang, South Korea, discussed the importance of characterising a new probiotic and understanding its mechanism of action to fulfil regulatory requirements. Benjamin Jensen, Department of Biomedical Sciences, University of Copenhagen, Denmark, stressed the importance of considering the gastrointestinal target and the diet and condition of the patient when developing probiotics, and Martin Haranta, PerBiotiX – Microbiome Solutions, Kysucké Nové Mesto, Slovakia, expanded on this subject, emphasising the promise that personalised and targeted modulation of gut microbiota holds for chronic disease. Sean Gibbons, Institute for Systems Biology, Seattle, Washington, USA, and University of Washington, Seattle, USA, described some of the current and emerging tools to support research into microbiota-based therapies and how these could be harnessed to achieve personalised medicine. Finally, links between the gut microbiome and the liver and central nervous system (CNS), and potential therapies to exploit these connections were illustrated by Peter Konturek, Thuringia-Clinic Saalfeld, Germany, and Gerard Clarke, University College Cork, Ireland.
Updates to the International Life Sciences Institute Europe Concise Monograph on Dietary Probiotics, Prebiotics and Gut Microbiota in Human Health
Arthur Ouwehand
Ouwehand emphasised that there is a need for reliable and objective information on what probiotics and prebiotics are, and what effects they can have on human health and disease.1 One of the aims of the ILSI is to address this need by publishing freely available, objective, scientifically sound, and concise monographs, written in language accessible to the interested layperson.2
In 2013, the European branch of ILSI (ILSI Europe) published a concise monograph on the topic of probiotics, prebiotics and the gut microbiota, which resulted from a collaboration between the Probiotic and Prebiotic Task Forces and external academic experts.3 Due to rapid advances in the understanding of these topics over the past 8 years, this monograph has been revised and republished this year, with the title ‘Dietary probiotics, prebiotics and the gut microbiota in human health’.2
Ouwehand presented the new aspects of the latest revision of the concise monograph, explaining that it now also includes sections covering:
The use of prebiotics to mimic the effect of human milk for infant formula: Oligosaccharides in breast milk have been shown to enhance the growth of bifidobacteria in the infant gut, which has led to the addition of prebiotics with a bifidogenic effect being increasingly added to infant formulae. Studies have shown that the addition of some prebiotics results in a gut microbiome, stool pH, and small-chain fatty acid (SCFA) pattern similar to breast-fed infants.
Prebiotics and probiotics in metabolic health, weight management, and food intake: Prebiotics have been associated with a reduction in food intake and decreasing fat mass, which can be beneficial in diabetes, though data is inconsistent in humans. Prebiotics have also been shown to increase SCFAs and modify bile acid profiles, as well as improving insulin sensitivity and glucose homeostasis. Some probiotics may have benefits in terms of weight management, possibly through their influence on satiety and hormones that control hunger.
Mental health and the gut–brain axis: Evidence suggests that intestinal microbes and their metabolites affect the brain, influencing behaviour. Early studies suggest that prebiotics and probiotics may have a beneficial effect on stress and anxiety.
The impact of prebiotics and probiotics on chronic inflammatory bowel disease (IBD): Research suggests that IBD may result from dysregulation of the mucosal immune response to the gut microbiota in genetically susceptible individuals, and that prebiotics and probiotics may have a positive impact on IBD risk and management.
The revised ILSI Europe concise monograph provides an update on the current understanding of the role that the intestinal microbiota play in health and disease, the proposed health benefits of probiotics and prebiotics, and an expanded definition of the term ‘prebiotic’ to include human milk oligosaccharides.2
Ouwehand concluded by stressing that although there are still gaps in understanding how probiotics and prebiotics work, the last few years have improved this knowledge considerably. He also emphasised that ILSI Europe is working on translating the concise monograph into other languages.
New Guidelines on the Use of Probiotics and Why Guidelines May Differ
Hania Szajewska
Szajewska highlighted some of the negative criticism directed towards probiotics in both scientific journals and the media, and explained that conflicting guidelines for the use of probiotics in healthcare are part of the problem.4 Guidelines in probiotics may differ because of inclusion/exclusion rules, the targeted population, the availability of probiotics, and the outcome measures considered.5
Szajewska summarised the latest recommendations for the use of probiotics from the European Society for Paediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN), which are currently under review for publication. She explained that the ESPGHAN working group restricted their recommendations to specific strains, or combinations of strains, of probiotics. In addition, recommendations were only made if at least two randomised controlled trials (RCT) were available for a particular strain. Szajewska also stressed that ≥80% agreement among experts in the ESPGHAN working group was required before any recommendation was made. Finally, the ESPGHAN guidelines were made available for public consultation over 16 days, and all comments were evaluated. The ESPGHAN guidelines are intended to help local policy makers to make decisions regarding the routine use of probiotics.6
Szajewska presented a recent RCT as an example to explain why guidelines can differ.7 This study assessed the effect of an eight-species probiotic on the risk of antibiotic-associated diarrhoea in children receiving a broad-spectrum antibiotic (n=3137 [NCT03334604]).8 Children were given either the probiotic or placebo for the duration of their antibiotic treatment and 7 days afterwards.7 The findings showed that the probiotic had no significant impact on antibiotic-associated diarrhoea (caused by Clostridoides difficile or of unexplained aetiology).7 However, probiotic treatment was associated with a significant reduction in relative risk of diarrhoea in general (regardless of aetiology) compared with placebo (relative risk: 0.65; p=0.02).7 Szajewska stressed that this shows the significant impact that definitions of diarrhoea have on outcomes in clinical trials, which affects guidelines that consider different outcome measures. This study represents the only RCT to investigate this eight-strain probiotic in paediatric antibiotic-associated diarrhoea. Therefore, it would not have been considered by the ESPGHAN working group. Other guidelines might take this RCT into account and provide recommendations based upon it, demonstrating the difference that inclusion criteria can make in clinical guidelines.
In terms of how clinicians can evaluate the differences between guidelines, Szajewska presented several possible approaches:
The ESPGHAN 2020 guidelines both recommended the use of Lactobacillus rhamnosus, Saccharomyces boulardii, and Lactobacillus reuteri probiotics for acute gastroenteritis. However, the American Gastroenterological Association (AGA) 2020 guidelines do not recommend the use of probiotics for this condition.9,10 These are clearly opposing recommendations, one conditional for and one conditional against. Szajewska explained that the interpretation of each of these recommendations is actually identical, and that different choices will be appropriate for different patients.9,10 Also, clinicians should help patients to make decisions on an individual basis that includes patient preference.
The ESPGHAN and AGA guidelines9,10 both recommend probiotics for the prevention of necrotising enterocolitis, yet the strains they recommend differ. Szajewska suggested that if there is any consensus between guidelines, such as the recommendation for L. rhamnosus in this case, then clinicians should use this strain if it is available in their country. If not, they should turn to a treatment that is recommended by at least one current guideline.
Guidelines may be unable to provide recommendations either for or against the use of probiotics, prebiotics, or synbiotics in a particular population, which means that the decision should be made between the physician and the patient to include their clinical, demographic, and family circumstances. For example, the European Academy of Allergy and Clinical Immunology (EAACI) guidelines for females who are pregnant or breastfeeding and infants indicates the options available to clinicians to help families to consider the pros and cons of different formulations.11
Szajewska concluded that there is no ‘one size fits all’ recommendation for probiotic use, and that probiotics should be considered alongside other available interventions to achieve the best outcome for each patient.
Probiotics on the Border Between Functional Food and Pharmabiotics
Sin-Hyeog Im
Dysregulation of the intestinal microbiome contributes to the development of immune disorders, neuronal disorders, and metabolic syndromes.12 Both microbes and their bioactive molecules are therefore candidates for microbiome-derived therapeutics.12 However, probiotics are not universally beneficial, and there have been cases where they were associated with harmful effects in at-risk groups.13 The U.S. Food and Drug Administration (FDA) and the European Directorate for the Quality of Medicines and healthcare (EDQM) now recognise probiotics as a new category of medicinal products termed ‘live biotherapeutic products’ (LBP).14 Specific guidelines are required for the use of LBPs, and regulatory approval of LBPs should require the identification of their effector molecules and their mechanism of action.14
Im explained that these requirements present challenges for the development of microbiome therapeutics, partly because of the difficulty in translating data from animals to humans due to the concept of the holobiont, which is the host genome and its associated microbiome.15 For example, there is a need to characterise LBPs in a similar way to drug candidates, evaluate them in a way that considers the immune and microbiome status of the patient, define its mechanism of action, and establish safety, efficacy, and quality control of the LBP.
Im described an innovative and recently developed pre-clinical model system designed to model the human immune system and microbiome.16,17 Avatiome™ (ImmunoBiome, Pohang, South Korea) can be used to develop novel drug candidates for microbiome-derived therapy.16,17 In this system, in vitro analysis is followed by ex vivo analysis and then by analysis of the microbiome and immune system in animal models, including specific pathogen-free mice, gnotobiotic mice, mono-associated with defined bacteria in germ-free mice, and a humanised mouse model.17,18 These assessments are followed by bioinformatic analysis, artificial intelligence analysis, clinical design, clinical indicator analysis, and new drug development target deduction analysis, to evaluate the human applicability of drug candidates.16,17
The Avatiome system has been used to identify bacterial strains with therapeutic potential as LBPs, IMB001 and IMB002, and their effect on cancer, autoimmunity, and autism spectrum disorder.16,17 By identifying the effector molecules from these bacteria, Im explained that it has been possible to define the underlying mechanisms of action. For example, IMB001 enhances the immunostimulatory effects of immune checkpoint inhibitors through rhamnose-rich heteropolysaccharides. Rhamnose-rich heteropolysaccharides are expressed on the bacterial surface and have roles in the activation of cytotoxic T cells and iron sequestration in the tumour microenvironment. IMB002 has anti-inflammatory properties through cell surface β-glucan/galactan polysaccharides, which induce the proliferation of regulatory T cells, and have been shown to suppress inflammation in mouse models of colitis.16,17 Therefore, Avatiome™ is currently being used to develop microbiome therapeutics based on these LBPs, aiming to improve anti-tumour immunity and reduce inflammation in IBD, in systemic lupus erythematosus, and also in autism spectrum disorder.16,17
Ecology and Physiology of the Small Intestine Versus Colon: Potential Consequences for Probiotic Administration?
Benjamin Jensen
In addition to directly influencing disease trajectory, gut bacteria and their secreted molecules can modify drugs and affect host cells, including their secretory products, such as bile acids.18 However, since much of our current understanding of the human gut microbiome comes from studying faecal samples, we may be missing host-microbe interactions that occur in metabolic and immunological niches in the small intestine.18
The small intestine and colon are physiologically quite different. Most simple nutrients are digested by host enzymes and absorbed in the small intestine, where the rapid transit time of luminal contents, along with high concentrations of bile acids and host-defence peptides, creates a hostile environment for bacteria. In contrast, the more favourable environment of the colon allows the microbiota to thrive, producing enzymes necessary for the fermentation of dietary fibre.19
Jensen emphasised the importance of considering the gastrointestinal target when developing probiotics. For example, while Crohn’s disease can affect both the small intestine and the colon, ulcerative colitis is restricted to the colon and rectum.20 Recent studies illustrate the importance of understanding interactions between the microbiome and the gut surface when developing probiotics. Firstly, Yu et al.21 found that colonic Paneth cell metaplasia is associated with increased colonic lysozyme production in patients with IBD. However, despite the protective anti-bacterial effect of lysozyme in the small intestine of healthy individuals, in mice with colitis, colonic lysozyme processing of mucolytic bacterial species such as Ruminococcus gnavus has been shown to exacerbate inflammation.21 Verma et al.22 showed that isolated cell surface β-glucan/galactan polysaccharides of Bifidobacterium bifidum induce regulatory T cells in the colon but not in the small intestine, indicating that bacterial isolates represent a potential alternative to live probiotic therapy for rebalancing colonic immunity and gut microbiota. Another option is to use lysates from non-commensal bacteria. Lysates of the non-commensal bacterium, Methylococcus capsulatus (Bath), which is found in soil, have recently been shown to induce regulatory T cells in the colon and small intestine of obese mice compared with a control diet.23 Jensen indicated that this approach may be relevant for the future treatment of a diverse spectrum of diseases, including food allergy (a small intestinal disease), ulcerative colitis (a large intestinal disease), and Crohn’s disease.18
It is also important to consider the effect of diet when developing microbiome-based therapies, and Jensen presented the example of two contradictory studies. Kovatcheva-Datchary et al.24 reported that an improved glucose metabolism following a high-fibre diet was associated with an enrichment of Prevotella copri in the gut microbiota of healthy subjects. Mice fed a high-fibre diet exhibited improved glucoregulatory capacity upon treatment with P. copri. Conversely, Pedersen et al.25 reported that P. copri was associated with increased serum levels of branched-chain amino acids characteristic of patients with insulin resistance. This study found that P. copri could induce insulin resistance in mice fed a low-fibre, high-fat diet, suggesting that diet per se can promote a dichotomous bacterial response in relation to human health. Another study that illustrates the importance of considering diet in conjunction with probiotics showed that in mouse models of obesity, high-fat diets based on different sources of fat, carbohydrates, and proteins produced differential responses to probiotics administered to induce weight loss (Larsen et al., unpublished data).
Finally, Jensen stressed that the individual circumstance and physical condition of each patient should be taken into consideration when developing or administering probiotics. For example, a recent clinical study identified selective plasma and microbial tissue signatures in age-, gender-, and weight-matched patients with morbid obesity, with differences between patients with and without Type 2 diabetes.26 These recent findings indicate that bacterial translocation may vary and depend on the health and disease status of each patient.27
Towards a Personalised and Targeted Microbiome Modulation
Martin Haranta
The composition of the gut microbiota is influenced by many factors, including age, health status, diet, genetics, and geographical region.26 Haranta stressed that while gut dysbiosis is associated with many diseases, the diversity, composition, and function of the gut microbiome can vary not only between diseases, but also among individuals suffering from the same disease. It is not yet clear whether gut dysbiosis is a cause or consequence of disease, but it is apparent that targeted gut microbiota modulation can alleviate symptoms, and will likely serve as adjuvant therapy for many diseases in the future.28,29
Haranta explained that novel approaches are needed to develop effective methods to modulate the microbiome for the prevention and therapy of chronic diseases. One potential approach is to personalise probiotic therapy, which should reduce the risks of treatment resulting in minimal benefit and/or adverse effects.28 A key challenge for personalisation is the selection of suitable biomarkers to direct the composition of a probiotic mixture. Personalised gut microbiota modulation is still in the early stages of research, and biomarkers that are currently used remain quite simple, such as the abundance of certain phyla or species in gut microbiota. However, Haranta emphasised that the metabolic activity of the microbiome can differ even while the composition remains the same, indicating that further research into metabolomics is needed to enable the effective personalised modulation of the gut microbiota.
A recent observational study was conducted to assess the application of personalised probiotics.30 Stool samples collected from volunteers (n=48) were analysed for microbiota composition, and this data plus a patient-reported medical history were used to prepare a personalised mixture of probiotics for each patient.30 A mixture of lyophilised probiotic cultures was prepared in capsules, with each single daily capsule containing 35 billion colony-forming units.30 The basis of the probiotic mixture was determined by the number and ratio of Lactobacilli and Bifidobacteria in the stool sample 30 For example, a low number of one type of bacteria resulted in the use of more of these bacteria in the probiotic mixture.30 Other factors used to prepare the personalised probiotic mixture included the ratio of Fermicutes and Bacteriodetes, which included a history of diarrhoea, antibiotic use, constipation, IBD, or allergy.30 Also, the BMI of the patients was included in the evaluation.31 After 3 months of probiotic administration, a second stool sample, clinical evaluation, and medical history were obtained from each patient.30
The results from this important study identified a statistically significant increase from baseline to month 3 in the mean number of Lactobacillus (0.01% versus 0.13%; p<0.0001) and Bifidobacterium (2.87% versus 4.08%; p<0.01) genera, and Actinobacteria phylum (4.84% to 7.18%; p<0.01), and in the total number of species in the faecal samples (799–1498; p<0.05). Haranta explained that these unanticipated increases in diversity illustrated the potential of this approach in improving gut microbiome health. The probiotic mixture was also shown to reduce self-reported diarrhoea to ≤2 defecations per day, or increase self-reported constipation to ≥1 defecation every other day, in more than half of patients studied.30
Haranta emphasised that personalised and targeted modulation of gut microbiota holds considerable promise as a strategy for preventing and treating chronic diseases. He envisages that this approach may be combined with auto-transplantation of ex vivo modulated gut microbiota in the future.
Tools to Leverage the Microbiome to Predict Personalised Responses to Dietary, Prebiotic, and Probiotic Interventions
Sean Gibbons
Gibbons explained that the interplay between a patients’ genome, microbiome, and environment (e.g., diet, exercise, medication) determines their phenotypic trajectory in terms of health and disease.31 This means that the composition of the gut microbiome can result in heterogeneous responses to interventions. There are various current and emerging tools to enable scientists to harness the microbiome for precision health, including in silico methods, in vitro methods, and in vivo methods.32
In Silico Approaches
Machine learning and mechanistic modelling have been investigated to predict personalised responses to interventions, often leveraging a deep phenotyping approach that includes the gut microbiota of the patient. For example, Zeevi et al.33 used a machine learning algorithm to predict postprandial glycaemic responses to non-standardised, real-life diet by analysis of meal composition (n=800). The algorithm was then used to design personalised dietary interventions which were predicted to be ‘good’ and ‘bad’ for each patient, depending on whether they resulted in low or high postprandial glycaemic responses. The ‘good’ dietary intervention significantly lowered postprandial glycaemic responses in 10 out of 12 participants and resulted in alterations to gut microbiota composition, compared with the ‘bad’ diets.33
In contrast to machine learning, mechanistic modelling does not require training, but an in-depth knowledge of the microbiome to generate novel hypotheses for causal mechanisms.34 A recent study applied MICOM, a customised metabolic model of the human gut microbiome, to undertake a metagenomics study of 186 people, including both healthy subjects and patients with diabetes.35 The results showed that individual bacterial genera maintained conserved niches across all participants, but that the production of SCFAs on the same exact background diet was more heterogenous.35
In Vitro Approaches
Gibbons described two in vitro approaches that can be used to assess personalised responses to microbiome-based interventions. The two approaches include culture-based systems, such as ex vivo faecal microbiota batch culture,36 or the continuous culture system, Simulator of the Human Intestinal Microbial Ecosystem (SHIME).37 However, although these systems are cost-effective and can maintain high throughput, Gibbons stressed that some organisms could be difficult to culture in this way, and these systems lack host tissue for the organisms to interact with. Therefore, to overcome this limitation, alternative models, such as the ‘gut-on-a-chip’ system, have been developed, whereby host cells and microbiota can be cultured together.38
In Vivo Approaches
Non-human in vivo approaches are usually closer to human models than in vitro systems, and permit experimental control over genetics and environmental exposures.31 Also, the availability of germ-free animals allows for causality testing.31 However, these systems are not biologically identical to humans,39 which limits the translational relevance of experimental findings. There are also ethical concerns inherent in using non-human animals in research.
Human in vivo studies are the approach most likely to produce findings that translate to the clinic. However, this approach lacks genetic and environmental control over the subjects, difficulty in obtaining some types of sample, and high cost. Gibbons posited that the crossover trial was one of the best methods to assess and personalised responses to dietary, prebiotic, and probiotic interventions.
Leveraging the Gut Microbiota to Predict Personalised Responses in the Clinic
The ultimate goal of high throughput design of personalised interventions to improve health and nutrition, represented in Figure 1, will be difficult to achieve. Gibbons explained that artificial intelligence platforms are needed to accomplish this goal.41
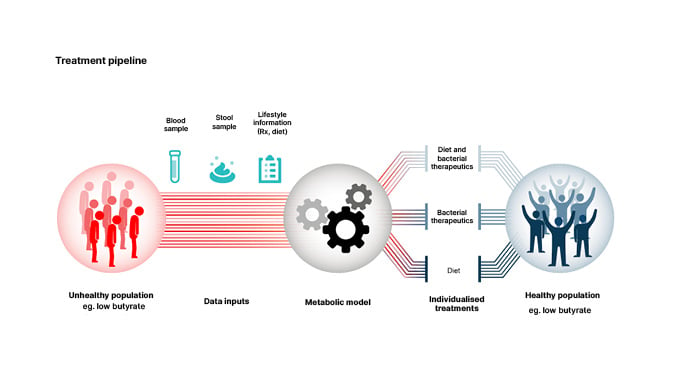
Figure 1: Personalised interventions for optimising gut microbiome health.
Reproduced from Wilmanski et al.40
One potential approach is to use a metabolic model such as MICOM35 to predict personalised engraftment probabilities for probiotics or pathogens and personalised microbial metabolite production profiles (e.g., Figure 1) in the context of a given microbiome composition and diet.
Many of the tools needed to achieve precision nutrition and healthcare have already been developed. Gibbons explained that the next step is to integrate these systems, optimise and refine precision models that target specific nutritional/clinical applications (e.g., glucose responses or SCFA production), and encourage their adoption by healthcare systems and healthcare workers.
The Gut-Liver Axis: From Bench to Bedside
Peter Konturek
Following exposure to gut-derived bacteria via the portal circulation, the liver activates innate and adaptive immune responses that can lead to hepatic injury.42,43 Several studies have demonstrated significant alterations in the gut microbiome and permeability of the intestinal barrier in chronic liver diseases, and microbiota-based strategies to manage these conditions can include diet, antibiotics, prebiotics, probiotics, and faecal microbiota transplantation (FMT).42,44
Konturek explained that pathogen-associated molecular patterns in bacteria and bacterial metabolites influence liver function in many different ways, including effects on inflammation, apoptosis, cytoprotection, regeneration, and bile acid composition.44
In non-alcoholic fatty liver disease (NAFLD), gut dysbiosis has been associated with disease progression.45 The key mechanisms involved in the impact of gut dysbiosis and the progression to non-alcoholic steatohepatitis are disruption of intestinal SCFAs, bile acids, and choline metabolic homeostasis, increase in lipopolysaccharides and endogenous alcohol, and activation of nucleotide-binding domain, leucine-rich-repeat containing family, pyrin domain-containing 3 and 6.46 Similarly, disruption of the gut-liver axis has been shown to contribute to the pathogenesis of metabolic dysfunction-associated fatty liver disease, including dysbiosis of the gut microbiota and bacterial overgrowth, thinning of the gut mucus layer, increased permeability of the gut epithelial barrier and vascular barrier, resulting in increased inflammation in the liver.47 Gut dysbiosis has also been linked to alcohol-related liver disease,48 primary sclerosing cholangitis,49 primary biliary cholangitis,50 autoimmune hepatitis,41 and liver cirrhosis.51
Probiotics, Prebiotics, or Synbiotics in Chronic Liver Disease
Manipulation of the gut microbiome by probiotics, prebiotics, or synbiotics has been shown to improve chronic liver disease.46 For example, a meta-analysis of four randomised trials involving NAFLD or non-alcoholic steatohepatitis patients found that probiotic therapy significantly decreased alanine transaminase, aspartate transaminase, total cholesterol, high-density lipoprotein, TNF-α, and insulin resistance (all p<0.05).52 Similarly, probiotics have been shown to significantly reduce the risk of hospitalisation for hepatic encephalopathy in patients with cirrhosis.53 Prebiotics such as fructo-oligosaccharides, galacto-oligosaccharides, and inulin have also been shown to benefit patients with liver disease, including significant decreases in alanine transaminase and aspartate transaminase. An RCT in NAFLD reported a significantly increased mean reduction in hepatic steatosis and fibrosis, and in serum inflammatory mediators, in patients receiving synbiotics compared with those receiving placebo.54
Faecal Microbiota Transplantation in Chronic Liver Disease
In a small RCT, 15 patients with NAFLD received an allogenic FMT from a healthy donor and six received an autologous FMT. Although there were no significant differences between groups in terms of insulin resistance or hepatic proton density fat fraction 6 weeks after FMT, patients with increased small intestinal permeability at baseline significantly reduced after allogenic compared with autologous FMT.55 In patients with severe alcoholic hepatitis presenting as acute-on-chronic liver failure (n=33), a single FMT from a healthy family member was associated with improved survival and improvements in clinical severity scores compared with standard of care.56 Finally, FMT compared with standard of care has been shown to increase microbiome diversity and improve cognition in patients with cirrhosis with recurrent hepatic encephalopathy.57
Konturek concluded that despite considerable research, the complex interactions between microbes and chronic liver diseases are not fully understood. Further studies are needed to develop novel therapies to treat these conditions.42
The Microbiota-Gut-Brain Axis: Therapeutic Targets
Gerard Clarke
Signalling pathways between the gut and brain are regulated at immunological, hormonal, and neural levels.58 These pathways are vital for homeostasis, maturation of the immune system, and CNS signalling (Figure 2).58
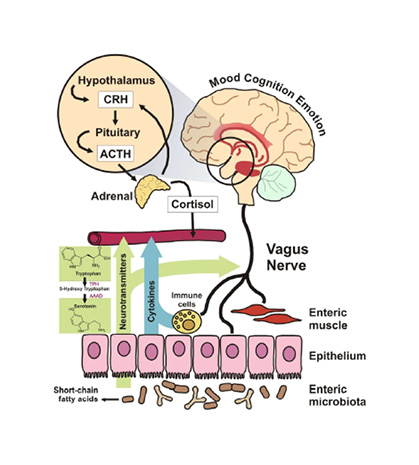
Figure 2: Proposed mechanisms of action through which microbiota can modulate signalling along the gut-brain axis.
Reproduced from Grenham et al.58
ACTH: adrenocorticotropic hormone; CRH: corticotrophin-releasing hormone.
Clarke explained that by manipulating the signalling pathways of the gut-brain axis, gut microbiota could modify brain function and behaviour, and research over the past 20 years has uncovered many of these host-microbe interactions.59
Research using animal models has revealed various ways microbiota affect the brain. For example, germ-free mice have an exaggerated stress response,60 and probiotics have been shown to reduce stress-induced corticosterone levels.61 Studies of brain structure in germ-free versus conventional mice suggests that microbiota are necessary for the normal development of the gross morphology and ultrastructure of the hippocampus and amygdala.62 Gut microbiota also play a role in serotonin metabolism, as male germ-free mice had higher concentrations of serotonin in the hippocampus, and higher serum concentrations of the serotonin precursor, tryptophan, compared with conventional mice.60 Interestingly, gut bacteria have been shown to secrete tryptamine, a serotonin-like molecule also formed from tryptophan, which acts through the 5-hydroxytryptamine-4 receptor in the colon to increase intestinal secretion.63 Finally, gut bacteria are known to produce SCFAs, and these metabolites have been shown to modulate microglia maturation in mice.64
Clinical research studies have shown reduced gut microbiome diversity and a link with reduced Prevotella and clinical depression.65 Microbial expression of 3-β-hydroxysteroid dehydrogenase has also been linked to changes in mental health.66 Furthermore, FMT from patients with depression to microbiota-deficient rats induced characteristic features of depression and alterations in tryptophan metabolism in the recipients.65 Finally, a meta-analysis of 34 case-control studies found that perturbations of gut microbiota, including depletion of anti-inflammatory, and enrichment of pro-inflammatory bacteria, were associated with depression, bipolar disorder, anxiety, and schizophrenia.67
Research into the potential of microbiome modulation as therapy for CNS disorders is still in its infancy,58 and Clarke presented some of the data that has accumulated to date. Bifidobacterium longum was shown to reduce stress and improve memory in healthy human volunteers,68,69 whereas L. rhamnosus failed to do so, despite promising animal studies to modulate stress or cognitive performance in healthy subjects.70 In another study, a diet that included polydextrose dietary fibre improved cognitive flexibility in conjunction with a significantly increased abundance of Ruminiclostridum 5 in the gut, compared with placebo.71
Psychobiotics and microbial metabolites are currently being investigated as therapies for psychiatric and neurodevelopmental disorders, as well as neurodegenerative disease. Clarke stressed that a better understanding of the microbiota-gut–brain axis will be critical to the success of these approaches.59