BACKGROUND AND AIMS
With the progress in upper gastrointestinal endoscopic technology, the usefulness of endoscopic early diagnosis and treatment of hypopharyngeal cancer has been reported.1 However, it is unfeasible to perform detailed pharyngeal observation requiring sedation for all subjects undergoing upper gastrointestinal endoscopy. There is an urgent need to establish a minimally invasive and simple evaluation method for high-risk patients with hypopharyngeal cancer. The aim of this study was to verify the diagnostic accuracy of superficial hypopharyngeal cancer by salivary DNA promotor methylation analysis.
MATERIALS AND METHODS
In total, 61 patients with superficial hypopharyngeal cancer who underwent endoscopic resection at Okayama University Hospital, Japan (derivation cohort), 23 patients at Hiroshima City Hospital, Japan (validation cohort), and 51 healthy controls (control cohort) were recruited. The authors assessed DNA methylation values of four genes (DCC, PTGDR1, EDNRB, and ECAD) in tissue and saliva samples by quantitative methylation-specific PCR for bisulfite converted DNA.
First, DNA was extracted from the superficial hypopharyngeal cancer area and the surrounding normal mucosal area in tissue samples resected at Okayama University Hospital. The accuracy of the discrimination between cancerous and normal mucosa was examined using DNA promoter methylation. Second, in a derivation study of saliva samples, DNA methylation values were analysed using the salivary DNA of the derivation cohort and control cohort. After establishing the cut-off value for differentiating between patients and healthy subjects, the diagnostic accuracy was evaluated. Third, as a validation study, the authors analysed DNA methylation levels in the salivary DNA of the validation cohort. They determined the detection rate in patients using the cut-off value calculated in the derivation study.
RESULTS
In the resected tissue samples, DCC (methylation value in normal mucosa: 3.94; in cancer: 12.10; p=0.003), EDNRB (11.84; 29.90; p=0.001), and ECAD (1.09; 4.73; p=0.043), showed significant differences in the amount of methylation between cancer and non-cancer site, except PTGDR1 (3.30; 4.33; p=0.47). When performing receiver operating characteristic analysis of salivary methylation for superficial pharyngeal cancer diagnosis in the derivation cohort, the area under the curve of DCC was 0.917 (95% confidence interval [CI]: 0.864–0.970), while the area under the curve of EDNRB and ECAD were 0.680 (95% CI: 0.576–0.784) and 0.639 (95% CI: 0.441–0.838), respectively (Figure 1).
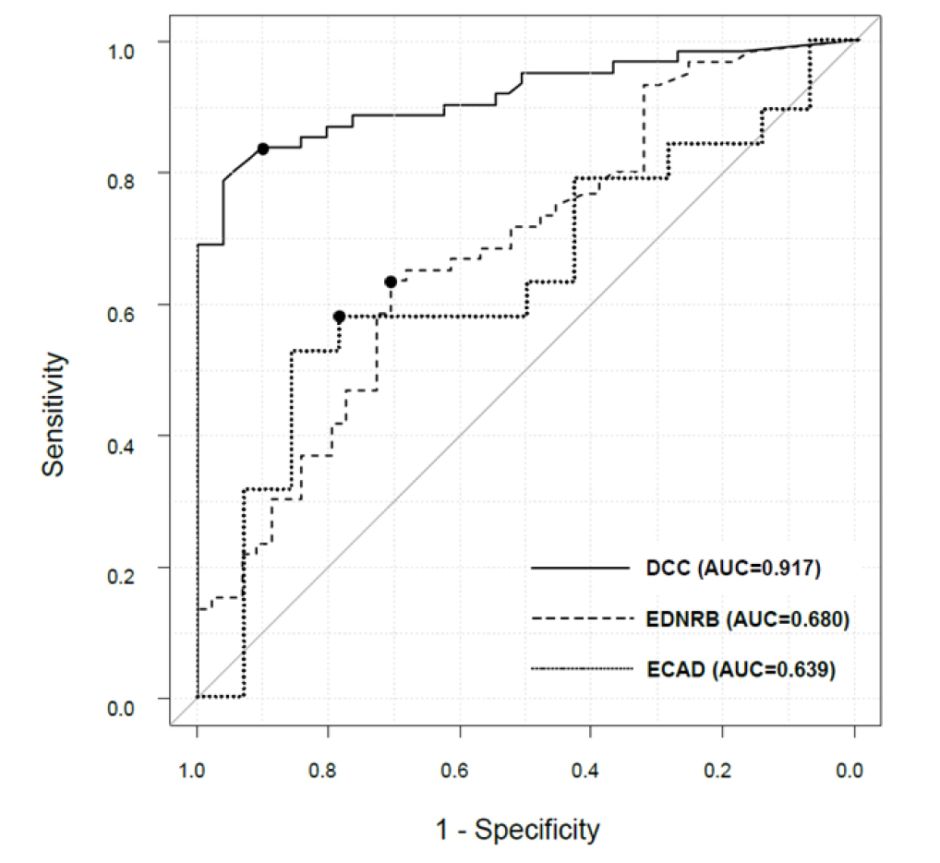
Figure 1: Comparison of the diagnostic accuracy for superficial hypopharyngeal cancer by the methylation analyses of the genes in the saliva.
The black points in each chart indicate the position of the cut-off value. The AUC of the ROC analysis of DCC, EDNRB, and ECAD for superficial hypopharyngeal cancer diagnosis were 0.917 (95% CI: 0.864–0.970), 0.680 (95% CI: 0.576–0.784), and 0.639 (95% CI: 0.441–0.838), respectively. The diagnostic accuracy of DCC methylation in saliva samples was significantly higher than that of EDNRB and ECAD methylation (p<0.001, log-rank test). The cut-off values for the normalised methylation value of DCC, EDNRB, and ECAD were ≥0.163, ≥0.256, and ≥0.655, respectively.
AUC: area under the curve; CI: confidence interval; ROC: receiver operating characteristic.
With the cut-off for the DCC methylation value in saliva samples setting at ≥0.163%, the sensitivity to detect hypopharyngeal cancer was 83.6% with a specificity of 90.2%.
Applying this cut-off value to 23 cases of the validation cohort, 18 cases (78.3%) of patients with superficial hypopharyngeal cancer were detectable.
CONCLUSION
In patients with hypopharyngeal cancer, salivary DNA methylation analysis of DCC showed high diagnostic accuracy even at the stage of superficial cancer. Because saliva can be collected easily and non-invasively, this method could be a useful tool for screening patients who are at high risk for hypopharyngeal cancer.