Abstract
Brain tumours are caused by the abnormal growth of cells in the brain. This occurs mainly due to genetic changes or exposure to X-ray radiation. When the tumours are detected early, they can be removed via surgery. The tumour can be removed through radiotherapy and chemotherapy if the removal of the tumour through surgery affects the survival rate. There are two main classifications of tumours: malignant or cancerous and benign or non-cancerous. Deep learning techniques are considered as they require more minimal human intervention than machine learning; they are built to accommodate huge amounts of unstructured data, while machine learning uses traditional algorithms. Though deep learning takes time to set up, the results are generated instantaneously. In this review, the authors focus on the various deep learning techniques and approaches that could detect brain tumours that were analysed and compared. The different types of deep learning approaches investigated are convolutional neural network (CNN), cascaded CNN (C-CNN), fully CNN and dual multiscale dilated fusion network, fully CNN and conditional random field, U-net convolutional network, fully automatic heterogeneous segmentation using support vector machine, residual neural network, and stacked denoising autoencoder for brain tumour segmentation and classification. After reviewing the algorithms, the authors have listed them based on their best accuracy (U-net convolutional network), dice score (residual neural network), and sensitivity score (cascaded CNN).
Key Points
1. AI technologies enable faster and more precise diagnostic capabilities by analysing large datasets from medical imaging and tests. Clinicians can use AI to complement their diagnostic expertise and improve patient outcomes.2. The adoption of AI in healthcare presents challenges around data privacy, ethical usage, and regulation. Clinicians must be mindful of these aspects to ensure AI is integrated responsibly and aligns with medical ethics.
3. AI is a tool designed to augment clinical decision-making, not replace it. Clinicians should focus on collaborating with AI systems, ensuring that human oversight and expertise remain central to patient care.
INTRODUCTION
A brain tumour develops as a result of abnormal cell growth in the brain. Tumours are classified as malignant or benign. Malignant tumours are cancerous, whereas benign tumours are non-cancerous. The other types of tumours are meningiomas, gliomas, and pituitary tumours. Meningiomas are mostly benign tumours originating from the arachnoid cap cells, representing 13–26% of all intracranial tumours. When the meningioma cannot be completely resected, radiosurgery is used to treat brain tumours.1 Gliomas are the most generally observed tumours, with different shapes and ambiguous boundaries, making them probably the hardest tumour to identify.2 Pituitary tumours are viewed as benign cancers, yet roughly 10% of them can have a forceful way of behaving, and once in a long while (0.2%) can introduce metastasis called pituitary carcinomas.3
The tumours show symptoms despite the size and part of the brain being affected. These include headaches, seizures, vomiting, and, in extreme cases, difficulty walking.4 Chemotherapy, radiation therapy, and surgery are used to treat brain tumours.5 Surgeries carry risks since the brain is a non-fungible organ. After surgery, medications are used to reduce the swelling around the tumour; seizures must be avoided, or another set of medications is needed.6 These treatments have a better success rate if tumours can be detected early and accurately. A few environmental factors that cause brain tumours are vinyl chloride or ionising radiation exposure. This leads to the mutation and deletion of tumour suppressor genes, causing brain tumours.7 Another factor is inheritance; conditions like neurofibromatosis Type 2 carry a high risk of developing brain tumours.8,9
Deep learning techniques are frequently used for image segmentation, classification, and optimisation in the medical industry in order to detect cancers precisely. There are different scanning techniques used to detect tumours; a few of them are CT, single photon emission CT (SPECT), PET, magnetic resonance spectroscopy (MRS), and MRI. CT has altered the course of symptomatic therapy since its invention in the 1970s. Increased radiation from patients is one of the major problems brought on by the increasing usage of CT.10 SPECT is a method generally utilised in nuclear medicine for imaging numerous organs, including the skeleton and heart, as well as for entire body imaging for the recognition of tumours.11 The advancements in PET are assessed with an accentuation on instrumentation for clinical PET imaging. PET provides good-quality images, high diagnostic accuracy, and short imaging protocols.12 The nuclear magnetic resonance principle underpins MRS. It can be used almost in any tissue of the body, but the brain is the major organ of interest in MRS. For biochemical characterisation, MRS provides an invasive diagnostic tool.13 This review uses an MRI scanning technique for comparison and analysis.
Brain MRI scans are used, and these include effective and quantitative analysis. MRI is frequently depicted as a protected methodology since it uses no ionising radiation.14 MRI is a sophisticated imaging strategy that has emerged as a clinical methodology in recent years.15 The process of image segmentation involves classification based on pixel-to-pixel techniques. There are many deep learning approaches that are used to detect these tumours such as convolutional neural network (CNN), contourlet convolutional neural networks (C-CNN), Fully Automatic Heterogeneous Segmentation using Support Vector Machine (FAHS-SVM), Fully Convolutional Neural Network and Dual Multiscale Dilated Fusion Network (FCNN and DMDF-Net), Residual Neural Network (ResNet), CNN (parametric optimisation approaches), CNN based Computer Aided Diagnostic (CAD) System approach, Cascaded CNN, FCNN and Conditional Random Field (CRF), Stacked Denoising Auto Encoder (SDAE), U-Net Convolutional Network (U-NET), and CNN (semi-supervised learning). All these approaches make use of MRI images as data input. Some pre-processing of raw MRI images is performed, such as removing unwanted parts of the image, enhancing contrast, etc., to improve the processing time and accuracy of algorithms. This review primarily focuses on identifying the best deep learning-based approach for segmenting and classifying brain tumours.
METHODS
The research articles were obtained from Scopus, Web of Science, and PubMed using the combined keyword phrase ‘Brain Tumor Segmentation and Classification’. Fifteen research articles on the topic of ‘Brain Tumor Segmentation and Classification’, published from the year 2017 onwards, were selected and used for this study. The research articles were studied to understand the type of deep learning approach used to detect the tumour in the brain. The uniqueness of each approach, the datasets used for the research, the type of tumour detected, limitations, and accuracy/scores were also checked.
Datasets
The following are the datasets used for brain tumour segmentation and classification. From a hospital in China, Nanfang, and Tianjing Medical University, a dataset has been collected from 233 patients. The dataset includes the years from 2005–2010. It contains 3,064 slices with 708 meningiomas, 1,426 gliomas, and 930 pituitary tumours in sagittal, coronal, and axial views. With cross-validation indices, 80% of images are employed for training and the rest for performance measurements.16 A dataset consisting of multi-spectral brain MRIs includes images of benign tumours, specifically meningiomas.17 A dataset tested from The Cancer Imaging Archive (TCIA) from Thomas Jefferson University, Philadelphia, Pennsylvania, USA, includes a total of 4,069 brain images, consisting of 3,081 non-healthy images (with abnormalities) and 988 healthy brain images, which were used for testing.18 And finally, a brain tumour dataset of 253 images, of which 98 were healthy and 155 were tumour-affected. About 80% of the images were used for training the model, while the rest were for testing purposes.19 The following experiments were conducted using the brain tumour segmentation challenge (BRATs) dataset from 2013–2020.
Experiments Conducted in BRATs 2013
This included T1, T2, fluid-attenuated inversion recovery (FLAIR) images of 30 patients. The tumour labels are annotated as (1) necrosis, (2) oedema, (3) non-enhancing tumour, (4) enhancing tumour, and 0 was denoted for normal tissue.20,21 This dataset consisted of 65 patients with glioma, including 14 low-grade gliomas (LGG) and 51 high-grade gliomas (HGG) from different centres: University of Bern, Switzerland; University of Debrecen, Hungary; Heidelberg University, Germany; and Massachusetts General Hospital, Boston, USA. The MRI scans included T2, T1, T1-c, FLAIR.22
Experiments Conducted in BRATs 2015
Training sets containing 220 patients with HGG and 54 with LGG were included. This involved all four MRI modalities. The labels were listed as follows: necrosis, oedema, non-enhancing tumour, and enhancing tumour.23 It included all modalities of MRI with an imaging size of 240x240x155. The modal images were linearly positioned to match the typical human brain.24 This included images from BRATs 2012, 2013, and TCIA. It consisted of 110 cases of unknown grades, 220 HGG, and 54 LGG for training.22 It also included images from medical image computing and computer-assisted intervention (MICCAI), which has data on patients with glioma. All MRI techniques were used, and they were classified as normal brain tissue, tumour, oedema, necrosis, or increased tumour.25
Experiment Conducted in BRATs 2016
It included 220 patients with HGG and 54 patients with LGG with 191 cases with unknown grades.22
Experiments Conducted in BRATs 2017
The experiments included 146 photos from patients with brain tumours and a validation set of 46 people who had the disease, whereas the testing sets are unknown. All the modalities of MRI were used.26 It included scans of 210 patients with HGG and 75 patients with LGG. Classifications such as a full tumour, oedema, enhancing tumour, necrosis, and non-enhancing tumour were given.27
Experiments Conducted on BRATs 2018
Patient data on histological subregions, aggressiveness, and prognosis were included, as well as multimodal MRI scans. The dimensions of the image are 240x240x150. Of 75 cases of LGG and 210 cases of HGG, 80% of it were for training, 10% for validation, and 10% for testing. It was grouped as necrosis, oedema, non-enhancing tumour, or enhancing tumour.6 It included 285 MRIs of patients with LGG or HGG. The images were acquired from 19 imaging centres. It was labelled as non-tumour, contrast-enhancing core, non-enhancing core, or oedema.28 A validation set of 66 patients with brain tumours and images from 191 patients with brain tumours were included, while the testing sets are unknown. All the modalities of MRI were used (FLAIR, T1ce, T1, and T2).26
Experiments Conducted on BRATs 2020
An experiment conducted on BRATs 2020 contains 370 images of HGG and LGG tumours. Following their use, all MRI modalities were classified into peritumoural oedema, necrotic/non-enhancing tumour, or enhancing tumour tissues.29
DISCUSSION
Convolutional Neural Network (Multiscale Approach)
A brain tumour segmentation and classification model using a deep CNN that includes a multiscale approach was used by Díaz-Pernas FJ et al.16 The method analyses MRI scans contained meningioma, glioma, and pituitary tumours from different viewpoints. The purpose was to develop this approach by using a T1-CE MRI scan dataset. Multi-pathway MRI scans were processed pixel by pixel by CNN architecture to cover the full image and separate the tumour-affected areas from the healthy ones. The size of the sliding window was about 65×65 pixels and 11×11 pixels. The feature maps were classified into large, medium, and small, and the classification of three types of tumours was hence defined. A few equations were used as part of the neural network training to get the results. The dice, sensitivity, and percentage tissue type agreement score pttas (score) were calculated and graphed into a histogram. The accurate score (pttas) was identified through the histogram. After the confusion matrix was calculated, the tumour classification accuracy was also calculated (0.973). Finally, a graph was draughted between the tumour classification and confidence threshold. This approach was compared with seven other deep learning approaches, and this was concluded to be the best among the others.16
Cascaded Convolutional Neural Network
Ranjbarzadeh R et al.6 considered a pre-processing approach to work with small parts of the image rather than the whole image. C-CNN incorporates both local and global characteristics into the two routes. The distance-wise attention mechanism was used to improve the accuracy of brain tumour segmentation, and it takes into account the effect of the tumour and the brain’s centre location within the model. The experiment was carried out using the BRATs 2018 dataset. The biological arrangement of the brain’s visual cortex serves as an inspiration for the CNN model’s structure.
For the experimental results, there are three criteria, namely HAUSDORFF99 for assessing the distance between the predicted and ground-truth region; sensitivity, which measures tumour pixels; and dice similarity, which computes the overlap between the ground truth and prediction. Comparisons were made with the baseline on BRATs 2018 dataset, and even though the proposed method had outstanding results, the algorithm was able to encounter tumours that were larger than one-third of the entire brain.6
Fully Automatic Heterogeneous Segmentation Using Support Vector Machine Approach
For segmenting brain tumours, FAHS-SVM was employed by Jia Z and Chen D.17 High-level homogeneity between the adjoining brain tissue’s structure and the segmented area made the segmentation functional. An extreme learning machine algorithm was used for getting regressions and classifications. The results showed that the accuracy was almost 98.51%. These experiments were conducted on a multi-spectral brain dataset. The following steps are involved in the architecture of the proposed method. One is pre-processing, which improves the MRI’s quality and makes it appropriate for subsequent processing. Second, skull stripping makes it possible to remove more brain tissues from brain pictures. Third, morphological operation and segmentation, which uses the wavelet transformation for efficient segmentation of MRI. The set of high-level visual details, such as contrast, form, colour, and texture, make up the feature extraction. The comparison was made between CNN, U-NET, and U-net with residual connections (Unet-res).17
Residual Neural Network Approach
ResNet, an automated method, was used to segment brain tumours by Shehab LH et al.23 This method relies on addition between the output layer and its input to provide greater accuracy and faster training processes. The approach was used to locate the whole tumour area, core tumour area, and enhancing tumour area from the BRATs 2015 dataset. Since the 3×3 convolutional kernels have a larger kernel and a similar response field, they were used in the model. The three stages of the ResNet approach that were proposed here are pre-processing, segmentation by ResNet, and post-processing. In the pre-processing stage, bias field distortion on four MRI sequences is fixed, and the generated patches are normalised. ResNet 50 was used for segmentation since it has a deeper layer and lesser parameters, so the training time would be faster. Finally, in the post-processing stage, FLAIR images were considered to locate the tumour. Evaluation metrics were used for testing the brain tumour segmentation. Comparing it with a few other methodologies, it was found that ResNet50 was the best, as the core accuracy was 0.84 and the highest among the others, and the computation time for the proposed methodology was faster (62 mins) compared to the other methodologies.23
Convolutional Neural Network (Parametric Optimisation Approach)
Nayak DR et al.19 used a special CNN architecture, the CNN model, together with a parametric optimisation strategy to assess brain tumour MRI. The model’s accuracy rate remains constant throughout Taguchi’s L9 design of experiment. The collection contained 253 medical photos of the brain, 98 of which were in a healthy state and 155 of which were images with tumours. The finest solutions to engineering challenges were produced using the forensic investigation algorithm, material creation algorithm, and sunflower optimisation algorithm, producing an even or random population. Using the brain tumour MRIs dataset and the suggested CNN model, the sunflower optimisation and material creation algorithms demonstrated excellent performance throughout the simulations.19
Convolutional Neural Network Approach
The segmentation of brain tumours was automated by Annmariya E et al.29 using a deep learning model. It automated the differentiation of brain tumours using several MRI modalities. The MR sequences were independently determined by single-channel input and multi-channel input. Experiments were run on the BRATs 2015 dataset of 220 MR images. The input image was 144x128x96 in size, and the HighRes3DNet architecture was used. The first seven levels of the 20-layer structure were derived from the input data using a 3x3x3 voxel convolutional kernel. The medium-level features from the input were encoded in the following seven layers, while high-level features were extracted in the last six levels using a dilated convolutional kernel. In single-channel models with quantitative evaluation, FLAIR sequence produced higher segmentation accuracy. The model using FLAIR and T2W inputs produced a 0.80±0.10 dice index through dual-channel models, demonstrating better performance. The FLAIR sequence was more accurate than other sequences at segmenting brain tumours, according to the results of segmenting single-channel sequences.29
From the newly designed CNN, useful features from multi-modality images are learnt to combine multi-modality information. This technique is applied to most classifications, and the accuracy is high too. This method uses multimodal and complementary data from BRATs 2013 T1, T1c, T2, and FLAIR images. Slices from the x, y, and z axils are chosen as inputs for each voxel in the 3D picture. This design converts tri-planar 2D CNNs from 3D CNN problems because it is more effective at lowering computing complexity. Dice ratio is used for evaluation to get segmentation accuracy. In comparison with Menze et al.30 and Bauer et al.,31 the proposed approach had better mean accuracy.20
Fully Convolutional Neural Network and Dual Multiscale Dilated Fusion Approach
FCNN & DMDF techniques were used to obtain the segmentation results with appearance and spatial consistency by Deng W et al.24 The Fisher vector encoding method was used to analyse the texture features, i.e., to change the rotation and scale in the texture image. The study experimented on the BRATs 2015 dataset with a 3D size of a modal MR image of 240x240x155. The similarity of the dice segmentation results of the brain tumour were analysed using the metrics coefficient, sensitivity, and positive predictive value. Compared with the other traditional segmentation methods, this approach had improved accuracy and stability, with an average dice index of 90.98%.24
Convolutional Neural Network Computer-Aided Diagnostic Approach
Arabahmadi M et al.21 adopted this method as its deep learning strategy since CNN (CAD) systems can help identify brain tumours from MR images. This entailed pre-processing, which included the region of interest segmentation (for brain tumours), enhancement, noise reduction, resizing, and skull stripping of MR images. The tests are performed on the BRATs 2013 dataset, which contains 50 photos of 30 patients and 30 locations labelled as necrosis, oedema, non-enhancing tumour, enhancing tumour, or everything else. Both local and global features were implemented using this technique. In contrast to the global features, which used 14×14 filters, the local features used 7×7 filters. The pace was raised by 40% by the convolutional layer, which was the last layer. The results demonstrated that deeper networks with larger patches outperform shallower networks, with dice scores of 0.88, 0.61, and 0.59 for patch sizes of 28×28, 12×12, and 5×5, respectively.21
Cascaded Convolutional Neural Network Approach
Wang G et al.26 proposed a cascaded CNN to segment brain tumours into hierarchical subregions. To increase segmentation accuracy, a 2.5D network and test-time augmentation were also applied. Experiments were conducted on the BRATs 2017 dataset. 2.5D anisotropy CNNs with a stack of slices as input with a large intra-slice receptive field and a small inter-slice receptive field have been proposed. By decomposing the 3D 3x3x3 convolutional kernel, the anisotropic receptive field was made. The kernel size of the intra-slice and inter-slice convolutional kernel are 3x3x1 and 1x1x3, respectively. This method was compared with 3D U-Net, with the whole tumour by W-Net, tumour core by T-Net, and enhancing tumour core by E-Net, and adapting W-Net without using the cascade of binary prediction. The results demonstrated that uncertainty estimation aids in both improving segmentation performance and spotting probable mis-segments.26
Fully Convolutional Neural Network and Conditional Random Field Approach
FCNN and CRF approaches were used by Zhao X et al.22 to obtain segmentation with appearance and spatial consistency. These are obtained in axial, coronal, and sagittal views, and the slice-by-slice method is used for the segmentation of brain images. BRATs 2013, 2015, and 2016 were used to locate glioma tumours in patients. Compared with other methods, the evaluation results of BRATs 2013 are top-ranked.22
Stacked Denoising Autoencoders Approach
In the study by Ding Y et al.,25 multimodal MRI brain tumour pictures were segmented using SDAEs. After training, the raw parameters that feed into the classification feed-forward neural network are obtained. Then post-processing is carried out. On BRATs 2015, experiments were done, and a preliminary dice score of 0.86 was obtained. Each layer in the architecture comprises several hidden layers, an encoder and a decoder. Input and hidden layers make up encoders, and the decoder gets the output. Normal brain tissue, tumour, oedema, necrosis, and enhanced tumour are the five outputs that make classification. For further optimisation, the gradient descent method is used to classify the centre of the image. The evaluation was on true positive, false positive, false negative, dice, and Jaccard coefficients, and while comparing with other methods, SDAE showed better segmentation results than the others.25
U-NET Approach
The usage of U-NET as a deep learning approach is discussed in the study by Cherguif H et al.27 Real photos from the BRATS 2017 datasets for medical image computing and computer-assisted interventions were used. Effective segmentation was offered with a dice similarity coefficient metric of 0.81805 and 0.8103 for the utilised dataset. The architecture was composed of three sections: the encoder, which reduces the feature maps from 240×240 to the bottleneck; the bottleneck, which compresses the feature maps to 15×15; and the decoder, which expands them back to 240×240. For ease of comparison, UNET-1 and UNET-2 are used instead. Additionally, when compared to other techniques, this one has a dice similarity coefficient (DSC) of 0.81 for brain segmentation and an accuracy of 0.99 for a whole tumour with core one.27
Convolutional Neural Network (Semi-Supervised Learning) Approach
Mlynarski P et al.28 introduced the idea of extending the segmentation networks with image-level classification. The model was trained for segmentation and classification tasks to annotate images. The experiments were conducted on the BRATs 2018 dataset with 281 multi-scan images. An additional branch, UNET is proposed to exploit information on annotated images. The feature map size is 101×101, from which the classification branch has to output two global classification scores; i.e., tumour absent and tumour present. Initially, the kernel sizes were reduced to 8×8 and mean pooling was used to avoid information loss and optimisation problems. Once the feature map was reduced from 64 to 32, the size was 11×11 of a fully connected layer of classification. In comparison, this approach outperforms three binary segmentation problems in a multiclass setting.28
It has been found that the CNN approach has been used often. CNN is traditionally used for image-based tasks. Since images are an array of numbers indicating colour intensity, they are suitable for image classification. Hence, the CNN algorithm is the most commonly used.32 The deep learning model that is better than CNN and found in the paper is SVM. Having huge datasets separates the boundaries of two classes and provides a warning.33 It can also perform in n-dimensional space. Table 1 shows the comparison of the different deep learning methods used for brain tumour segmentation and classification with respect to accuracy, dice score, and sensitivity score. Comparing the research papers based on the accuracies and scores, the highest accuracy is found in U-NET, the highest dice score in ResNet, and the highest sensitivity score in C-CNN. Hence, each algorithm has advantages and disadvantages, displaying different scores and accuracies.
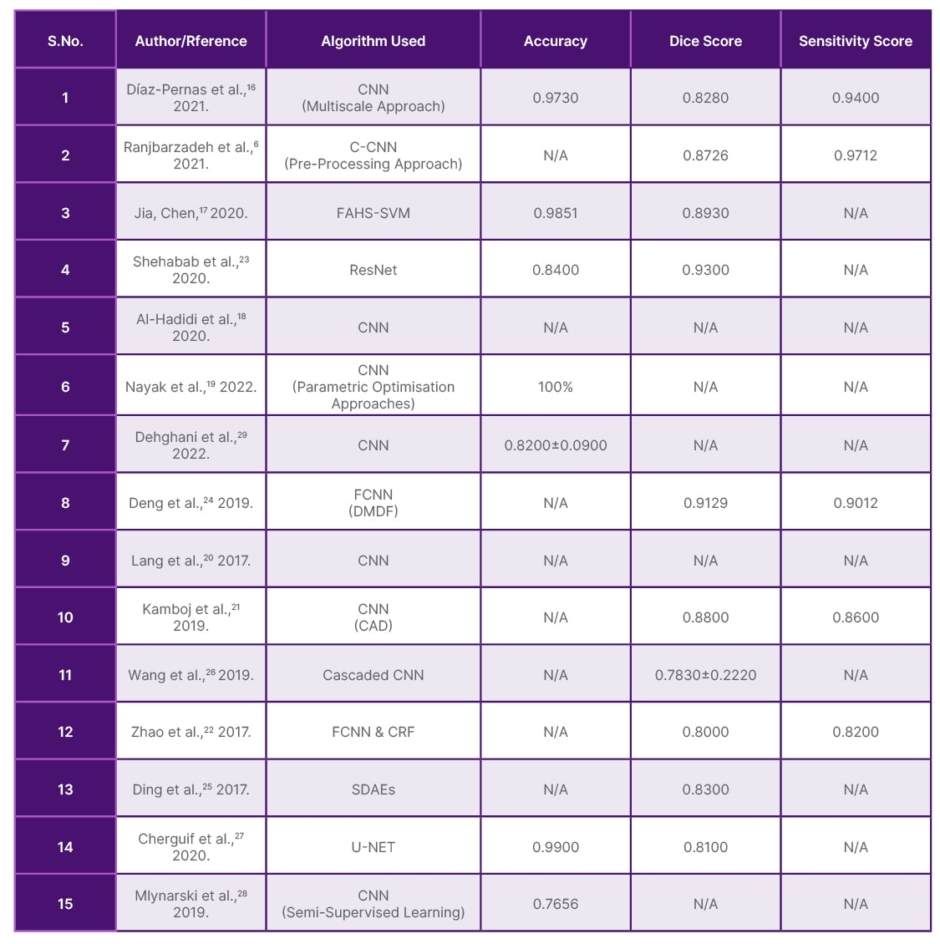
Table 1: Comparison of the different deep learning methods used for brain tumour segmentation and classification with respect to accuracy, dice score, and sensitivity score.
CAD: computer aided diagnostic; CNN: convolutional neural network; C-CNN: contourlet convolutional neural networks; CRF: conditional random field; FAHS-SVM: fully automatic heterogeneous segmentation using support vector machine; FCNN and DMDF-Net: fully convolutional neural network and dual multiscale dilated fusion network; ResNet: residual neural network; SDAE: stacked denoising auto encoder; U-NET: U-net convolutional network.
CONCLUSION
The advantage of using deep learning is that it provides high-quality accuracy. It is concluded that the U-NET algorithm has the highest accuracy compared with the others (0.99). Almost every article includes dice and sensitivity scores that check the performance of the image and how a positive value can be generated correctly. Taking this into consideration, for the dice score, ResNet approach has the highest value (0.93), and for the sensitivity score, C-CNN, which includes the pre-processing approach, has the highest value of 0.97. Future studies will focus on enhancing these findings and utilising deeper architectures to boost output segmentation performance. Methods for segmenting brain tumours are anticipated to be powered by weak, unsupervised training with fewer labels. For successful brain tumour segmentation networks, the authors anticipate combining tumour degrees such as Grade 1, 2, 3, and 4, where Grade 1 has less aggressive behaviour and tumour morphology, which records the type of tumour that has developed and how it behaves with neural architecture search algorithms. The growing role of AI in healthcare represents a paradigm shift, promising to revolutionise clinical practice, medical research, and patient care. AI’s potential to analyse vast amounts of data quickly and accurately can significantly reduce the workload of clinicians, allowing more time for patient interaction and personalised care. Nevertheless, the impact of AI will largely depend on how well it integrates with existing healthcare systems, its acceptance by medical professionals, and its ability to enhance rather than replace human judgement. Continuous education and training for healthcare professionals on AI technologies will be crucial in ensuring its safe and effective implementation.