Abstract
Introduction: The most common cutaneous fungal infections are caused by dermatophyte fungi such as Microsporum, Trichophyton, and Epidermophyton. In this study, the epidemiologic trends and the predominant organisms causing dermatophytosis in Urmia, Northwest Iran, were identified.
Aims and objectives: To get better perception of dermatophyte distribution in Northwest Iran, the authors studied the identification of isolated dermatophytes from human specimens by using a fast and cheap molecular method: PCR-based restriction fragment length polymorphism (PCR-RFLP). The authors also aimed to use this method in for rapid and reliable identification of medically important and common dermatophytes at the level of species.
Methods: The study samples were collected from clinically suspected cutaneous lesions. All the specimens were transported to Medical Mycology Center, Urmia Medical Sciences University (UMS), Iran. First of all, a conventional diagnosis was carried out, which included microscopic examination and culture of sabouraud dextrose agar medium with antibiotics: chloramphenicol and cycloheximide. All the dermatophyte isolates were then identified at the level of species by the molecular method of PCR-RFLP.
Results: From the tested 357 clinical specimens, 30 dermatophytic isolates were identified. The percentage rate of dermatophyte species were Trichophyton mentagrophytes (36%), Microsporum canis (32%), Microsporum gypseum (16%), Trichophyton rubrum (4%), and Epidermophyton floccosum (12%).
Conclusion: By using of PCR-RFLP, a fast and reliable identification of these species is possible. This molecular method provided an opportunity for dermatophyte identification at the species level.
INTRODUCTION
Dermatophytes are the keratinophilic moulds living on the superficial layer of human and animal skin and are transmitted by direct and indirect contact with infecting debris or soil.1-4 These fungi are not able to cause serious and fulminate infections. The diseases caused by dermatophytes may have important clinical consequences, including secondary bacterial infections, remedial failures, and mental difficulties.5 For the best choices of antifungal drugs or treatment protocols, it the reliable identification of the species level is necessary as some dermatophyte species such as Trichophyton rubrum are usually resistant to routine treatments. A correct and rapid identification of dermatophytes at the species level helps to improve the diagnosis of dermatophytic infections6 and control environmental and animal sources of infection, resulting in the development of preventive strategies.7
Routinely used characteristics for identifying of dermatophytes are clinical symptoms, culture parameters, microscopic features, and physiological examinations. However, the differentiation of dermatophytes is, in some cases, hard and confusing because of overlapping morphologic features, polymorphism, and shifting formation.8 In fact, the classic identification of isolates using morphologic features has been complicated by their overlapping characteristics, variability, and pleomorphism. Mating as a means of identification is not always practical as a result of the need to keep a library of opposite mating types for each species. Furthermore, many of the anamorphic species lack a teleomorph. A variety of chemotaxonomic methods have been developed to bypass the traditional methods of identification and to determine relationships between various species.9
During last few decades, many researchers have tried to design new molecular methods for fast identification of dermatophyte fungi at the species level in clinical specimens and cultures.10-11 Alternative molecular methods to the identification of dermatophyte fungi have been used through techniques such as arbitrarily primed PCR,12 random amplified polymorphic DNA analysis,13,14 and restriction analysis of mitochondrial DNA15 and recombinant DNA (rDNA),16,17 which are generally adequate for various species.
The authors studied the identification of isolated dermatophytes from human specimens by using the fast and cheap molecular method, PCR-based restriction fragment length polymorphism (PCR-RFLP) to get better perception of dermatophyte distribution in Northwest Iran. They also aimed to use this method in for rapid and reliable identification of medically important and common dermatophytes at the level of species.
METHODS
The authors’ cases were selected among patients with the clinical manifestation, suspected to be dermatophytosis. For 7 years, starting in October 2011, 357 clinical specimens were collected (100 samples taken during the Year 1). The specimens were taken from the scalp, body, palm, foot, and nails by scrapping the skin and hair lesions at the dermatology clinic of Imam Khomeini University Hospital, Urmia, Iran, and transported to the Medical Mycology Center, Urmia Medical Sciences (UMS) University, Iran. All clinical specimens were obtained from cases applied for routine mycological examinations; therefore, it was not necessary to prepare a letter of consent for the authors’ cases due to the provisions of the Committee of Ethics, and the authors also got approval from the Deputy of Research, UMS University. A direct microscopic examination was carried out on the specimens using potassium hydroxide 10–20%, and the wet mounts to detect the pathogenic forms of dermatophyte, including septate mycelia and arthroconidia.
Also, basic culture medias such as sabouraud glucose agar 2% and sabouraud agar, with chloramphenicol 50 mg/L and cycloheximide 500 mg/L, were used for the detection of dermatophytes in the specimens. Then, morphologic identification of dermatophytes at species level was carried out based on the macroscopic and microscopic characteristics. The macroscopic features, including colony size, topography and textures, production of pigment, and fruiting bodies, were considered18 as well as the microscopic characteristics, including shape and size of macroconidia or microconidia conidia, mycelia, and other characteristics.
DNA Extraction
Dermatophytic mycelia for the DNA extraction harvested from the 48–72 hours growth in sabouraud glucose broth. A manual DNA extraction using 0.4 mm glass beads and phenol–chloroform was performed. The lysis solution included: 1 mM ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate, 100 mM sodium chloride, 10 mM tris-hydroxymethil aminomethane hydrochloride (Tris-HCl), and 2% Triton X-100, mixed in distilled water.16
PCR for Identification
The PCR profile included 5 µL of the DNA template in a total volume of 50 µL, containing a PCR buffer (20 mM Tris-HCl at pH 8), 50 mM potassium chloride, and 0.1 mM of each primer. The authors used rDNA universal primers for the amplification of the internal transcribed spacer (ITS) regions (Primer F :5’- TCC GTA GGT GAA CCT GCG G – 3’; and Primer R: 5’- TCC TCC GCT TAT TGAT TAT GC – 3’) and 1.5 U of Taq polymerase DNA (Mirhendi Molecular Biology Lab, Tehran University of Medical Sciences [TUMS], Iran).16 The reactions performed in a thermocycler (XP Cycler, BIOER, Hangzhou, China). Thermal programme included an initial DNA denaturation at 95 °C for 5 minutes, followed by 30 cycles of denaturation at 95 °C for 30 seconds, annealing at 55 °C for 30 seconds, and extension at 72 °C for 1 min, with a final extension at 72 °C for 5 min. The PCR products were subjected to a 1.5% agarose gel electrophoresis and were documented using a UV documentation system (Syngene, Cambridge, UK).
Digestion of PCR Products
The restriction enzyme MvaI was used in RFLP.16 For each reaction, 13 µl of PCR product was directly digested by 5 U (0.5 µl) of the restriction enzyme in 1.5 µl of the enzyme buffer, at 37 °C for 180 min. Digested PCR products were subjected to 2% agarose gel electrophoresis. The identification based on the differential patterns among the medically important dermatophytes at the level of species (Table 1).
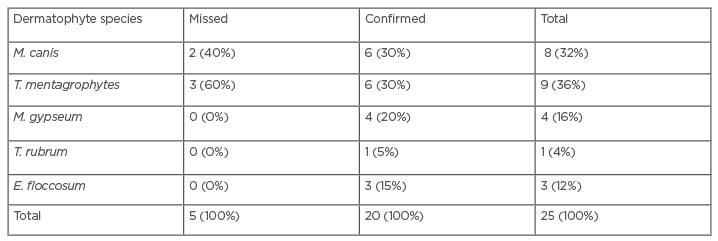
Table 1: Frequency and percentage of dermatophytes species identified by the PCR-restriction fragment length polymorphism method.
E. floccosum: Epidermophyton floccosum; M. canis: Microsporum canis; M. gypseum: Microsporum gypseum; T. mentagrophytes: Trichophyton mentagrophytes; T. rubum: Trichophyton rubrum.
RESULTS
As expected, amplification of rDNA’s ITS regions resulted in a PCR pattern with similar electrophoretic bands in size (600–700 bp), which distinguished no dermatophytes in this study (Figure 1). Digestion of the PCR products with MvaI in PCR-RFLP made a differential electrophoretic pattern that some tested dermatophyte species, including Trichophyton mentagrophytes, Microsporum canis, Microsporum gypseum, T. rubrum, and Epidermophyton floccosum were identified (Figure 1). The morphologic identification, confirmed by PCR-RFLP, include some macroscopic and microscopic features that are shown in the figures.
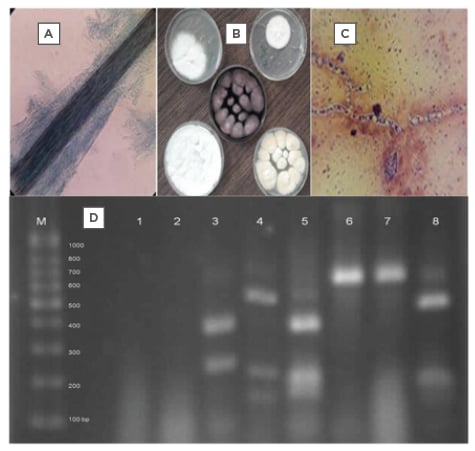
Figure 1A–D: The pictures A to C show the microscopic feature of a hair infection, some dermatophyte colonies and the microscopic picture of a dermatophytic mycelium (respectively), and picture D shows PCR-RFLP pattern of some identified dermatophytes in Lanes: 3, 5, 4, 8 and not digested DNA bands 6, 7 of the studied dermatophyte species.
Among all the studied cases, 25 (7%) were identified with a dermatophyte infection. The findings of PCR-RFLP confirmed five dermatophytic species including: T. mentagrophytes (36%), M. canis (32%), M. gypseum (16%), T. rubrum (4%), and E. floccosum (12%); although, two cases of M. canis and three of T. mentagrophytis were missed (Table 1). Among all the isolated dermatophytes, the T. mentagrophytes complex and M. canis were the most frequent species (Table 2). The most common sites encountered by dermatophytes were the scalp (skin and hair), nail, body, and palm, and the most frequent infections were tinea capitis and tinea unguium. Among all dermatophyte species, M. canis, M. gypseum, and the T. mentagrophytes complex were the most isolated, and Trichophyton schoenleinii was identified by PCR-RFLP as the exceptional case in this study.
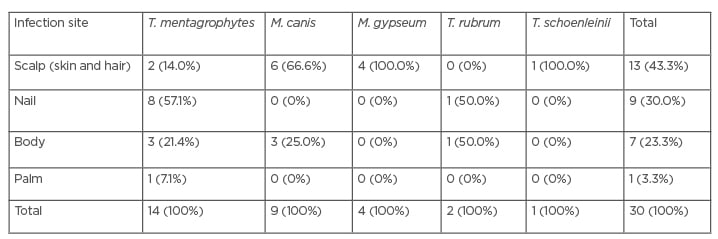
Table 2: Data of identification by the conventional method based on the involved site.
M. canis: Microsporum canis; M. gypseum: Microsporum gypseum; T. mentagrophytes: Trichophyton mentagrophytes; T. rubum: Trichophyton rubrum; T. schoenleinii; Trichophyton schoenleinii.
DISCUSSION
Conventional (phenotypic) identification of dermatophyte fungi is problematic due to a lack of stable characteristics distinguishing between isolates. Most T. rubrum strains show uniformity in both microscopically and colonial appearance, although variations in colony morphology do exist.19 Likewise, in some instances, the causative dermatophyte fails to produce any obvious reproductive structure in culture (termed sterile mycelia), which makes it impossible for ultimate definitive diagnosis.
Many typical isolates of common dermatophytes can be identified directly from primary isolation media, particularly sabouraud glucose agar and potato glucose or potato flakes agar. Identification characters include colony pigmentation, texture, and growth rate, and distinctive morphological structures such as microconidia, macroconidia, spirals, pectinate branches, pedicels, and nodular organs.20 In spite of its some disadvantages such as expensive material, including PCR kits and restriction enzymes, and the limited potency of identification for most species, PCR-RFLP prepares a differential pattern for the identification of dermatophytes to the species level in a rapid and reliable manner.
In the authors’ study, use of PCR-RFLP provided the authors with a simple and rapid diagnostic method compared with the conventional culture and microscopy. However, a reliable statistical comparison between the two methods used need much more identified cases from each dermatophyte species. The use of PCR-RFLP can be of great use; however, when it is not possible to use it for the above specified reactions, the classical method can be still valid and advisable for identifying species with well-characterised morphological aspects.
In the study by Kamiya et al.,21 identification was obtained from the PCR and PCR-RFLP targeting the DNA Type II topoisomerase gene and using some restriction enzymes in all cases. Also, Ganlin et al.22 identified six common dermatophytes by using PCR-RFLP targeting the Type II topoisomerase gene. All six dermatophytes were identified to species level.22
In the present study, the use of PCR-RFLP with the single restriction enzyme MvaI, according to the previous studies,23 enabled the identification of most of the Aspergillus species, which was proved in this study by using the morphologic method. There have been several restriction enzymes for the digestion in PCR-RFLP to better identify species, and the newer ones could have been used but in the present study, MvaI was selected to compare the data with the results of similar works. This is a well-known restriction enzyme for dermatophytes identification. All tested dermatophytes were identified at species level and no obvious difference found in terms of identification among the species patterns. Some more studies have also confirmed the present molecular findings: Mirzahoseini et al.24 studied the application of PCR-RFLP by using different restriction enzymes, including MvaI, HinfI, and HaeIII for the differentiation of isolated dermatophytes at the genus or species level.23
However, there were some exceptions in the present findings of molecular study, including M. canis, M. gypseum, and T. schoenleinii, which did not match with the other methods. In fact, 30 dermatophyte isolates (8.5%), recovered in the culture, were identified and confirmed by a molecular test based on the rDNA ITS regions. Molecular identification of the isolated dermatophytes provided reliable information about the frequency of dermatophytic infections in Northwest Iran. With the ignorance of the mentioned exceptions, M. canis, T. mentagrophytes, M. gypseum, T. rubrum, and E. floccosum were the most frequent isolated dermatophytes in Northwest Iranian cases in the present molecular study. Other Iranian surveys have presented similar results, as shown in studies by Mirzahoseini et al.24 and Zamani23, where T. mentagrophytes, T. rubrum, T. verrucosum, M. canis, and E. floccosum are the main isolated dermatophyte species.
An Indian epidemiologic survey reported predominant dermatophytes including T. rubrum, T. violaceum, T. mentagrophytes, and E. floccosum. In contrast to the present findings in Iran, the major isolates of scalp and body of the Indian study were T. violaceum and T. rubrum, respectively. Also, in a Japanese study, six dermatophyte species of T. rubrum, T. mentagrophytes, Trichophyton tonsurans, M. canis, M. gypseum, and E. floccosum were obtained from 305 patients with tinea. M. canis seems to be prevalent and a more common cause of dermatophytosis than other dermatophytes agents.21 It is believed that the distribution pattern of dermatophyte species in Asia follows a general incidence with some differences.
Furthermore, according to Glanin et al.,22T. rubrum, T. mentagrophytes, Trichophyton verrucosum, M. canis, M. gypseum, and E. floccosum represented the main causes of human dermatomycosis, which were the most frequently isolated species, in the dermatology department of University Hospital Graz, Austria. This point was also in agreement with the present results.
CONCLUSION
By using molecular method in the present study, a fast and reliable identification of medically important dermatophytes at species level is possible. To make the best drug choice for treatment, it is necessary the identity the dermatophyte fungi at the species level.






