Abstract
Atopic dermatitis (AD) is one of the most common non-infectious diseases in the world. For over two decades there has been considerable mobilisation to create a robust framework to address this global problem (the International Study of Asthma and Allergies in Childhood [ISAAC] consortium). However, information about Sub-Saharan Africa remains sparse, likely reflecting the increased focus placed on infectious diseases. However, this region harbours the greatest environmental and genetic diversity and thus offers enormous potential for understanding the differential environmental impact on human populations predisposed to allergic diseases. Moreover, it is increasingly clear that many pathologies share the same genetic determinants and this spans both non-infectious and infectious diseases. In this review, we discuss the comparative genetics of the allergic diseases and then expand into infectious diseases, notably malaria. We discuss the considerable overlap in the identified genetic determinants of AD and malaria and develop a hypothesis based on the importance of saliva from mosquito bites, arguably the most prevalent allergen in the region. Following the completion of the first phase of the African Genome project, we stress the significance of more focus on allergic diseases in the region, which will certainly generate an abundance of novel insight into the environmental and genetic determinants of allergy and may also contribute to our understanding of arthropod-borne infectious diseases.
INTRODUCTION
Atopic dermatitis (AD) is a common chronic inflammatory skin disorder that is often associated, either sequentially or concomitantly, with asthma and allergic rhinoconjunctivitis. The global significance of these non-communicable allergic diseases led to the establishment of the International Study of Asthma and Allergies in Childhood (ISAAC) consortium in the 1990s.1 The aim of ISAAC was to establish baseline rates of asthma, rhinitis, and eczema to enable global comparisons, assessment of future trends, and to provide a framework for aetiological research into lifestyle, environmental, genetic, and medical care factors affecting these diseases.2 However, at its inception (Phase 1), only three Sub-Saharan countries (Nigeria, Kenya, and Ethiopia) were included, likely because of the general focus on infectious diseases that impose the major health burden in this region. In Phases 2 and 3, several other countries joined and yet information on the allergy status remains sparse. Point prevalent studies were carried out in 12 Sub-Saharan countries, almost all in children aged 13–14 years old and in urban settings. Rates ranged from 4.7% in the Sudan to 19% in Ethiopia, with a mean of 14.5%.3,4 Only two studies have however, examined AD trends over time (Phase 1 in 1995 and Phase 3 in 2001–2). In Nigeria there was an increase in lifetime prevalence of itchy rash (7.7% to 10.2%) in children 6–7 years old but a strong decrease (26.1% to 18%) in the 13–14 year olds. Physician-diagnosed AD decreased significantly in both age groups.5 By contrast, the Kenyan study on 13–14 year olds in rural cohorts revealed a similar increase in AD to that found in the most studied African countries,6 Morocco and South Africa;7 lifetime itchy rash increased from 23.8% to 28.5% and physician-diagnosed AD from 13.9% to 28.5%.
The data on African populations is thus sparse and needs to be improved, not only for the specific public health burden that allergy may impose on the region, but also because studies in Africa may be pertinent for allergy worldwide. AD is very prevalent in African-American children,8 highlighting ethnic differential susceptibility. The human genome was shaped by the selective forces present prior to the early human migrations out of Africa, when infectious diseases were likely of significant importance. Such selection of strong pro-inflammatory responses by infectious diseases may have negative consequences in the form of allergic diseases worldwide today.9 The development of AD has been associated with genetic polymorphisms, skin barrier dysfunction, environmental exposures, and host immune dysregulation. The pathophysiology of AD may be associated with a child’s sensitisation to specific environmental or food allergens in association with skin barrier dysfunction. Africa is an important region to study complex diseases such as AD, because of its high genetic diversity, large variation in climate, and environmental conditions.10 The importance of both human genetic and environmental determinants of AD makes the dearth of information from Sub-Saharan Africa all the more frustrating. Following the completion of the first detailed characterisation of the African genome, it is now possible to benefit from particularities of the African genome and environment to address genetic and environmental factors that contribute to complex multifactorial diseases.11 Sub-Saharan Africa has a very particular environment and one that needs special attention, largely because of the high incidence of infectious diseases. As we will discuss here, the likelihood that there are shared human genetic determinants underlying infectious and non-infectious diseases should generate an increased capacity to identify genes of interest. Furthermore, infectious agents represent a facet of the environment that is rarely considered, likely because of the relatively low prevalence in societies where the majority of allergic disease research is carried out.
COMPARATIVE GENETICS OF ATOPIC DERMATITIS AND ALLERGIC DISEASES
Genome-Wide Association Studies
It is widely recognised that common multifactorial diseases are caused by multiple genetic and environmental factors and interactions among all these factors. With the development of genotyping technologies, genome-wide association studies (GWAS) have become the method of choice to identify complex disease associated genes using single nucleotide polymorphisms (SNPs) as biomarkers.12 One of the interesting features of GWAS is that the same loci have been found to be associated with several diseases (e.g. cancers, cardiovascular diseases, autoimmune diseases), suggesting that genes with a pleiotropic effect may be more frequent than anticipated and may play a key role in basic pathophysiological mechanisms underlying a number of diseases. AD is often associated with asthma and allergic rhinitis, and it is likely that these three diseases share common genetic and environmental determinants.13 The identification of pleiotropic genes that are likely to influence master regulators of biological processes is therefore of major importance. Studying together diseases that are suspected to share common genetic determinants can facilitate the characterisation of such genes.
There have been several GWAS studies of AD in European, Japanese, and Chinese populations.14-16 A total of 19 susceptibility genes have been identified, suggesting that AD is influenced by genes involved in epidermal barrier functions (e.g. filaggrin [FLG]) and innate and adaptive immunity amongst others (notably, interleukin [IL]-1 signalling and nerve growth factor signalling).17 The adaptive immune response in AD is associated with increased expression of the T helper 2 (Th2) cell cytokines, IL-4 and IL-13.18 Recent studies have shown a significantly greater number of IL-4, IL-5, and IL-13 messenger RNA (mRNA)-expressing cells in acute skin lesions, and an increased number of Th2 cells expressing IL-4 and IL-13 mRNA in unaffected skin of AD patients.19 Allergen-specific immunoglobulin (Ig)E plays a crucial role in the pathogenesis of allergic diseases by binding allergens and initiating immunological responses. Approximately 80% of patients with AD have elevated serum IgE and/or immediate skin test reactivity to allergens.18 There is a strong genetic contribution to the variability of the total IgE level. Several GWAS for total IgE levels have identified two associated susceptibility loci, 1q23 and 5q31, in European and mixed populations including African-American and Hispanic individuals.20-22 In particular, a functional SNP in 1q23 significantly influenced the cell surface expression of Fc fragment of IgE, high-affinity receptor (FCεR1A) on basophils, and the regulatory mechanism of FCεR1A expression via GATA2.20
Recent GWAS have revealed a number of susceptibility loci shared by several allergic diseases and phenotypes for bronchial asthma, allergic rhinitis, the number of eosinophils, allergic sensitisation, and total IgE levels. Moreover, there are significant genome-wide susceptibility regions overlapping AD and other allergy-related phenotypes. Of particular interest is the susceptibility loci 5q31.1 that contains RAD50 and IL-13, amongst others.23-25 The IL-4, IL-13, STAT6 pathway has been reported to be associated with IgE and asthma and the IL-13 gene is the only one found significantly associated with both asthma and IgE by the GABRIEL consortium GWAS.26 The association of STAT6 with IgE reported by a previous GWAS was replicated by the GABRIEL consortium.20,26
Filaggrin and Thymic Stromal Lymphopoietin
FLG plays an important role in the skin’s barrier function and FLG loss-of-function mutations have been associated with an increased risk of developing persistent AD.27 FLG binds to and condenses the keratin cytoskeleton to form tight bundles, flattening and strengthening the cells to create a strong barrier. Subsequent proteolysis leads to further degradation into hygroscopic amino acids, which constitute one element of natural moisturising factor. A defect in the production of FLG creates a more porous skin surface, resulting in epicutaneous sensitisation to environmental allergens and activating the host immune system, thereby resulting in local inflammation, pruritus, and visible skin lesions. FLG loss-of-function mutations are more commonly found in individuals of European and Asian ancestry compared with those of African ancestry.28 Indeed, of the four most common European mutations (R501X, 2282del4, R2447X, and S3247X), at least one was found in 27.5% of whites compared with 5.8% of African-Americans.29
In addition to FLG, the thymic stromal lymphopoietin (TSLP) gene was found to be located adjacent to a region (5q11.1) associated with AD. TSLP is a cytokine produced by epithelial cells (such as keratinocytes and bronchial epithelial cells) and has an important role in conditioning dendritic cell (DC) maturation. TSLP-activated DCs induce inflammatory Th2 cells that produce the classical Th2 cytokines IL-4, IL-5, and IL-13, in addition to large amounts of tumour necrosis factor (TNF). Interestingly, mast cells activated by IgE binding to the high- affinity IgE receptor (FcεRI) expressed high levels of TSLP.
TSLP promotes Th2 cell responses associated with immunity to some helminth parasites and the pathogenesis of a number of inflammatory diseases such as AD. Indeed, increased expression of TSLP has been strongly associated with AD as well as other allergic diseases, including asthma and allergic rhinitis.30-32 Although FLG protein contributes to the skin barrier, TSLP expression occurs after antigen sensitisation through a disrupted skin barrier and subsequently promotes the immune responses resulting in inflammation that leads to AD. In AD, epithelial cells markedly increase TSLP expression in response to inflammation, leading to macrophage activation, DC maturation, induction of inflammatory Th2 cells, and eventual chemoattraction of a suite of innate immune cells, such as eosinophils, neutrophils, and mast cells, with resulting pathological effects. Genetic variants resulting in diminished TSLP activity were protective against persistent AD even in individuals with the FLG loss-of-function mutation.32,33 Therefore, TSLP is a master regulator of allergic inflammation in the skin and can significantly influence the AD phenotype.
INFECTIOUS DISEASES AND ALLERGIC RISK
There are many examples of infectious agents that impact upon allergic risk. Severe respiratory syncytial virus infection in infants increases the risk of allergic rhinoconjunctivitis and allergic asthma.34 Measles,35 hepatitis A, and tuberculosis seemingly reduce atopy,36 whereas HIV infection is associated with increased risk of asthma and AD.37 Helminth infections are considered to reduce clinical expression of an atopic tendency.38 The relationship of Staphylococcus aureus and patients with AD is unusual in that S. aureus colonisation is both a cause and a consequence of allergic skin inflammation.39 The allergic skin inflammation associated with AD leads to increased colonisation by S. aureus notably because of skin barrier dysfunction and ineffective innate immune responses. S. aureus in turn produces exotoxins (superantigens) that can penetrate the skin barrier and contribute to the persistence and exacerbation of allergic skin inflammation.40 Of particular interest are pathogens that are transmitted by haematophagous arthropods (mosquitoes, sandflies, ticks) because of the allergenic nature of their saliva.
Lessons from Malaria
Although the human genetic susceptibility to malaria has classically focussed on the haemoglobinopathies (sickle cell, beta and alpha thalassaemia), there is an unusual relationship between allergy and the mosquito-borne protozoan parasite, Plasmodium falciparum, the aetiological agent of lethal tertian human malaria. Several lines of evidence support the concept that susceptibility to malaria and atopy may be related to the same immunological defect and/or influence one another.
Impact of allergy on malaria infection outcome
At the phenotypic level, there is evidence that allergy impacts upon risk of malaria. In Ethiopia, atopic children had a higher prevalence of malaria attacks,41 while in Tanzania maternal malaria had a protective effect on wheezing in 4-year-old children.42 AD, and to a lesser extent asthma, were found to increase the risk of persistent clinical malaria episodes, suggesting interference with the developmental of clinical immunity to the parasite.43 Finally, there is some evidence that IgE levels are elevated during malaria infections and that the mosquitoes as well as the parasites contribute to this state and thus may exacerbate the immunological environment.44,45 Notably, individuals classified as having moderate or severe symptoms of AD (assessed using the positive and negative predictive values of the ISAAC questionnaire diagnosis criteria developed for sub-tropical countries) had significantly higher specific IgE levels against Culex, Aedes, and Anopheles salivary gland extract.43,45
Role of immunoglobulin E
Several findings suggest that IgE could play a detrimental role during malaria disease development. This was supported by data showing that IgE levels were much reduced amongst patients with uncomplicated malaria in comparison with those suffering from severe malaria.46 Furthermore, immunohistological studies on brain sections revealed the presence of IgE deposits in brain microvessels and on infected erythrocytes from cerebral malaria patients as well as in placentas infected with P. falciparum.47 Various immune complexes, which consist of either IgE and antigen aggregates or IgE with IgG anti-IgE, could bind to Fc receptors expressed on monocytes that become activated, giving rise to TNF-a secretion.46 In addition, IgE levels were found to be higher in cerebral P. falciparum malaria when compared with uncomplicated malaria.48
Shared genetics
A mouse model (NC/Jic) for human atopic disease was found to be susceptible to murine malaria and a major quantitative trait locus (derm1) for atopic disease mapped close to the region controlling parasitaemia (char1 or pymr) on mouse chromosome 9.49,50 Genome scan linkage studies revealed four regions linked to malaria phenotypes. 5p15–p13 and 13q13–q22 were found to be associated with clinical malaria and 5q31–q33 and 12q21–q23 with parasite density.51 While these regions are extensive and contain many putative candidate genes, it is remarkable that all four regions overlap with those that have been previously identified to be involved in asthma/atopy and especially IgE levels,52,53 suggesting that common mechanisms may be involved between both pathogenic mechanisms. Moreover, a logarithm of the odds (LOD) score >1 was found using both analytical methods for the region containing the high-affinity receptor for IgE (FcεR1B on 11q12.1). The regions 13q13–q22 and 12q21–q23 contain genes known to increase total serum IgE levels, namely PHF1153 and STAT6 genes.54 The Stat6 protein plays a central role in exerting IL-4 mediated biological responses. The IL-4 gene is located on 5q31 together with important cytokines (IL-13, IL-5, IL-9), a region found to be linked to parasite density in this and previous studies and consistently reported to be linked to asthma-related phenotypes.55 The cluster of cytokines on 5q31, namely IL-4 and IL-13, are major cytokines promoting the differentiation of Th2 cells that are involved in the allergic response. Genetic variants in IL-4 and IL-13 genes on 5q31 and STAT6 on 12q21 have been consistently found associated with total IgE levels and this is confirmed by GWAS of this phenotype.20,56
Of particular interest is the 5p13 linkage region; this includes several genes involved in innate immunity and notably the interleukin-7 receptor (IL-7R). IL-7R plays a crucial role in signal transduction of TSLP. TSLP receptor chain alone binds to TSLP at low affinity. A combination of TSLP receptor and IL-7R-a chain results in high-affinity binding. Fine mapping of this 5p region using Illumina® GoldenGate revealed an association with IL-7R at a LOD score of ~5 (unpublished data).
MOSQUITO BITES, ATOPIC DERMATITIS, AND MALARIA: AN UNDERLYING MECHANISM
It is recognised that the type of immune balance driven by the parasite operates at a very early stage post parasite delivery. The response of sentinel cells, such as DCs, thus determines the evolution of the immune response and can lead to protection, tolerance, or immunopathology. Mosquito saliva contains pharmacologically active proteins and peptides57 which provoke a localised allergic reaction in the skin, and injection of saliva into the skin during a mosquito bite induces the production of IgE and IgG antibodies as well as dermal hypersensitivity reactions.58 This suggests that the saliva can orientate the immune response towards a Th2 profile.
The combination of this information leads us to propose a mechanism underlying the observed shared associations between AD and malaria (Figure 1). Mosquito bites will lead to activation of keratinocytes resulting in localised production of TSLP that subsequently orientates immature DCs to a Th2 profile. These in turn induce inflammatory Th2 cells that produce associated Th2 cytokines, which result in differentiation of B cells that generate IgE. Cross-linking of IgE on the high affinity receptors on mast cells causes their activation and release of both TSLP and histamine. Increased levels of histamine in plasma and tissue, derived from basophils and mast cells, notably following stimulation by IgE through the high affinity receptor FcεR1, are associated with the severity of disease in humans infected with P. falciparum and in animal malaria models.59 The release of TSLP by mast cells will in turn stimulate Th2 DC orientation and so forth. Perturbation of the Th1/Th2 balance will certainly impact upon the long-term development of immunity to malaria parasites and the short-term response to infection. Furthermore, the itch and scratch cycle characteristic of AD will be exacerbated by mosquito bites and saliva, which is a highly prevalent ‘environmental’ allergen in Sub-Saharan Africa and elsewhere. Mosquito bites will thus likely increase AD in individuals genetically predisposed and with resultant increased severity of malaria disease. Whilst the same relationship would be predicted for P. falciparum endemic settings out of Africa, it may not be the case for the other major malaria parasite, spp. Plasmodium vivax, which induces a very different immunological response. It is, however, still unclear whether the parasites themselves play an active role in such immune deviation, exacerbate AD in turn and the extent to which the parasites themselves benefit from the increased severity of disease associated with such an inflammatory terrain: the production of specialised parasites stages, necessary for transmission from human to mosquito, was reduced in individuals with elevated IgE.45 Further research on the physiological link between malaria parasite infection outcome and AD and other IgE-associated pathologies would be invaluable. Underlying details notwithstanding, it is promising to note that certain antihistamines have been effective against malaria and the current anti-malarial of choice, artesunate, against asthma.60,61 It remains to be seen whether next-generation allergic disease treatments (IL-4/IL-13 agonists) also have beneficial action against malaria parasites.
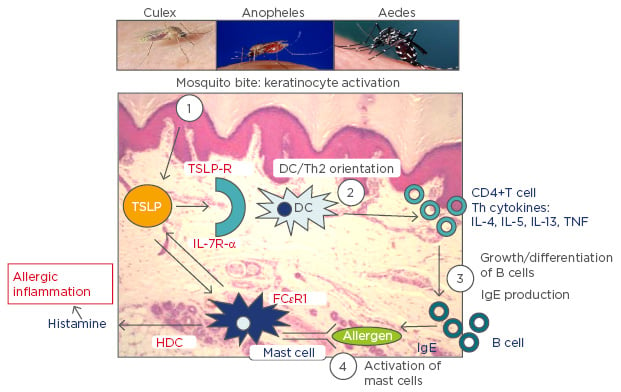
Figure 1: A schematic representation of the proposed key role played by mosquito bites in allergic inflammation.
TSLP: thymic stromal lymphopoietin; TSLP-R: thymic stromal lymphopoietin receptor; IL: interleukin; IL-7R-α: interleukin-7 receptor alpha; DC: dendritic cell; HDC: histidine decarboxylase; TNF: tumour necrosis factor; IgE: immunoglobulin E; Th2: T helper Type 2; FcεRI: high-affinity IgE receptor.
CONCLUDING REMARKS
The underlying causes of AD are multifactorial and yet it has been recognised for some time that environmental factors play an important role and increase or decrease risk in individuals with a genetic predisposition. Sub-Saharan Africa has received relatively little attention and yet represents a region that is rich in environmental and genetic diversity. A major challenge in diagnosis of AD is the plethora of different aetiologies that share similar symptoms. HIV/AIDS associated cutaneous symptoms in particular, may pose a significant challenge for AD differential diagnosis in Africa.8 Further focus on allergic diseases in this region would be of immense value to our understanding of the genetic and environmental basis to allergic diseases and will likely contribute to our understanding of infectious diseases and most notably those transmitted by biting arthropods.






