BACKGROUND AND AIMS
In early life, the immune and respiratory system are functionally immature, presenting a window of susceptibility in which a subset of infants are hypersusceptible to acute viral bronchiolitis (AVB) and at high-risk of frequenthospitalisations,1,2 for reasons incompletely understood. AVB is characterised by asthma-like symptoms, and clinical presentation is variable and may reflect a heterogeneous syndrome/subphenotypes.3 Importantly, susceptible infants are more likely to develop chronic pulmonary disease later in life, including persistent asthma.4 Studies in infants with AVB have been limited and mainly restricted to circulating blood-derived cells in infants aged <24 months.5-7 Thus, systems-level studies extending to nasal tissues with accompanying personalised immune response profiling are urgently required to advance the understanding of the precise underlying mechanisms of AVB, and to drive development of more precisely targeted therapeutics. The study approach taken in this study8 (Figure 1) consisted of a comparison of AVB-associated expression profiles in affected infants (aged <18 months) versus older children (>18 months–5 years), encompassing both circulating peripheral blood mononuclear cell (PBMC) and nasal mucosa, the site of primary viral infection. The aim of this study was to characterise the cellular and molecular mechanisms underlying severe respiratory infections in infants and children with AVB.
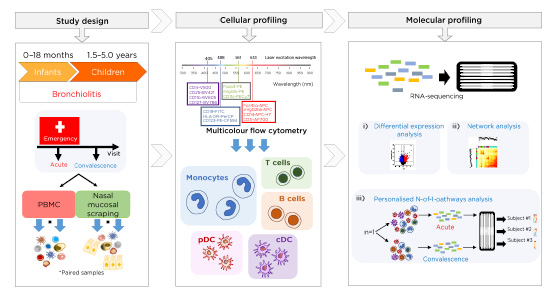
Figure 1: Study design and sample collection at the acute and convalescent visit of infants (aged <18 months) and children (aged >18 months – 5 years) with acute viral bronchiolitis.
Cellular (multicolour flow cytometry) and molecular transcriptomic (RNA-sequencing) profiling was carried out on cells obtained from peripheral blood and nasal scrapings. Analyses included differential expression analysis, network analysis, and novel personalised immune response profiling using N-of-1-pathways analysis.
cDC: conventional dendritic cells; PBMC: peripheral blood mononuclear cell; pDC: plasmacytoid dendritic cells.
METHODS
PBMC and nasal mucosal scrapings were obtained from infants (aged <18 months) and children (aged >18 months–5 years) during severe AVB and postconvalescence. Immune response patterns were profiled in a paired design following a battery of analyses ([hlFigure 1[/hl]): multiplexed plasma cytokines, viral diagnostics, flow cytometry, and transcriptomics (RNA-sequencing). Immune profiling firstly consisted of group-level systems analyses employing upstream regulator and coexpression network analysis. Secondly, potential heterogeneity among individual subjects was identified with personalised immune response profiling using N-of-1-pathways analysis.9
RESULTS
Group-level analyses demonstrated that infant PBMC responses were dominated by monocyte-associated hyperupregulated Type 1 IFN signalling/proinflammatory pathways (drivers: TNF, IL-6, TREM1, IL-1B), while in children a combination of inflammation (drivers: PTGER2, IL-6) plus growth/repair/remodelling pathways (drivers: ERBB2, TGFB1, AREG, HGF) coupled with Th2- and natural-killer-cell signalling pathways were upregulated. Potential confounders of molecular signatures, such as steroid usage and variations in underlying viral pathogens, were excluded as contributors to age-related differences between infants and children.
In nasal mucosal tissues, Type 1–3 IFN signatures were qualitatively comparable in infants and children; however, the magnitude of upregulation was higher in infants (range: 6–48-fold) than children (range: 5–17-fold). The most intense response profiles were observed in infants manifesting febrile symptoms.10 Personalised immune response profiling employing N-of-1-pathways analysis confirmed the upregulation of innate immunity in infants and natural-killer-cell networks in children, and additionally unmasked AVB response subphenotypes that were independent of chronological age.
CONCLUSIONS
A defining immunologic characteristic of AVB-susceptible infants was dysregulated expression of IFN-dependent pathways.8 Moreover, febrile infants showed uniquely complex immunoinflammatory responses during AVB,10 and a subset of children also demonstrated this IFN-hyperupregulated immunophenotype in peripheral blood. A subcluster of subjects manifesting susceptibility to severe respiratory viral infection in early life are characterised by delayed immune development trajectories, i.e., slow kinetics of postnatal maturation of innate immune competence.
CLINICAL IMPLICATION
Personalised immune response profiling uncovered covert intrasubject variation in immunoinflammatory phenotypes, a pattern which was concealed in ‘averaged’ expression profiles amongst this age group. Moving forward, personalised transcriptomics may be a key tool to identify risk-associated immunophenotypes and better inform the selection of appropriate targeted treatments.






