BACKGROUND
Enolases represent a unique family of allergens as they are not only found in all three major kingdoms (animals, plants, and fungi), but can further cause respiratory, food, and contact allergies.1 The ubiquitous occurrence and high homology suggest potential cross-reactivity between enolases.1 Although cross-reactivity between enolases from closely related species has been shown, it is not known whether IgE-reactivity to enolases from distantly related species occurs due to cross-reactivity or co-sensitisation.1-3 The purpose of this study was to analyse the cross-reactive potential of enolases from various allergen sources to gain a better understanding of the proteins’ pan-allergenic character.
METHODS
Enolases from different fungal species (Alternaria alternata, Aspergillus fumigatus, Penicillium chrysogenum, Cladosporium herbarum, Paecilomyces variotii, and Candida albicans) and animal-derived foods (chicken, pork, salmon, and trout), known as important respiratory and food allergen sources, were recombinantly produced in Escherichia coli. The molecules’ IgE-reactivity and cross-reactivity were investigated by ELISAs, immunoblots, and inhibition experiments with sera from polysensitised patients.
RESULTS
Enolases from 10 different sources were successfully produced in recombinant form and were analysed regarding their IgE-reactivity. Among them were also three enolases that have not yet been described as allergens, namely enolases from Penicillium chrysogenum (rPen c 6), pork (rSus s 6), and trout (rOnc m 6). Not only the seven enolases previously identified as allergens, but also the three novel enolases were recognised by patients’ IgE antibodies, suggesting the allergenic potential of these enolases (Figure 1A). Analysis of the IgE-reactivity patterns of 10 polysensitised patients to the recombinant enolases showed that patients always reacted with enolases from multiple sources, but they showed different sensitisation profiles (Figure 1B). All patients reacted to at least one fungal enolase, and two of them reacted only to fungal enolases. In addition, all the other eight patients recognised at least one meat enolase, and five of the eight patients furthermore reacted with at least one fish enolase.
IgE inhibition experiments revealed strong IgE cross-reactivity between enolases from closely related sources, since, for example, pre-incubation of the serum from Patient 1 with recombinant A. alternata enolase (rAlt a 6) led to complete inhibition of IgE-binding to the ELISA-plate coated enolases (Figure 1C). Patients’ IgE-binding to enolases was only partially reduced by enolases from distantly related sources. For example, pre-incubation of the serum from Patient 1 with the mold enolase rAlt a 6 led to a partial inhibition of IgE-binding to the yeast C. albicans enolase (rCand a 6; Figure 1C). Moreover, no cross-reactivity was yet observed between enolases from species belonging to different kingdoms (Figure 1D), as exemplified by the IgE-binding to pork enolase (rSus s 6) that could not be reduced by pre-incubation of the serum of Patient 3 with rAlt a 6. These data suggest that despite the high homology of enolases (60–93%), no cross-reactive IgE-epitopes are shared between enolases from distantly related allergen sources.
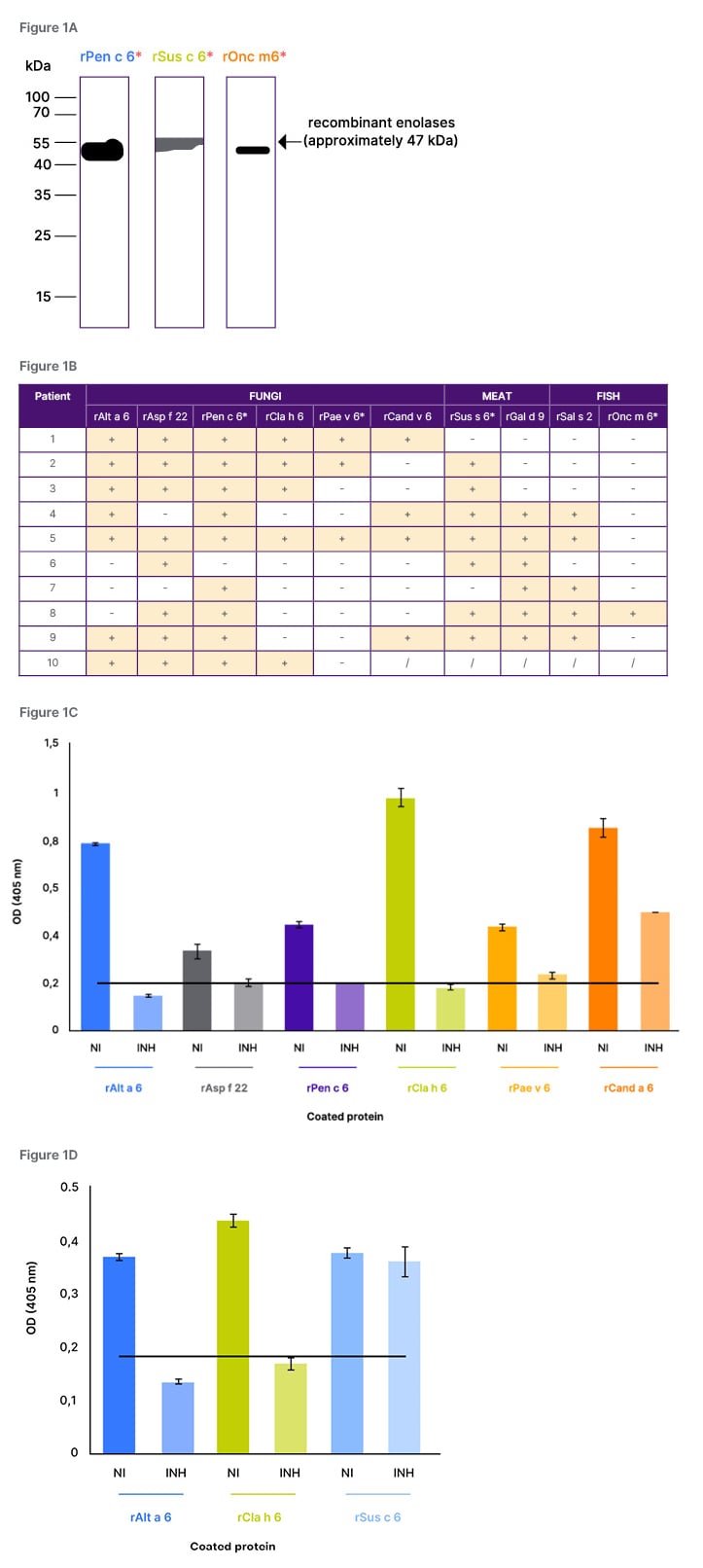
Figure 1: IgE-reactivity of polysensitised patients to recombinant enolases and IgE cross-reactivity.
A) IgE-reactivity of polysensitised patients to recombinant enolases from chrysogenum (rPen c 6), pork (rSus s 6), and trout (rOnc m 6) tested by immunoblot (*designated names given by the authors). B) Summary of IgE-reactivity patterns of 10 polysensitised patients to recombinant enolases from fungi and animals, tested by ELISAs. C/D) IgE-binding of the serum from Patient 1 (C) or Patient 3 (D), pre-incubated with rAlt a 6 to different recombinant enolases, tested by IgE inhibition ELISA.
INH: inhibited; NI: not inhibited; +: positive reason; -: no reaction; /: not tested; rAlt a 6: A. alternata enolase.
CONCLUSION
IgE antibodies from polysensitised patients recognised enolases from different allergen sources. Also, enolases from P. chrysogenum, pork, and trout were identified as potential allergens. IgE cross-reactivity was only seen between enolases from closely related species, whereas enolases from distantly related organisms lacked cross-reactivity. This suggests that IgE-reactivity might be caused by independent co-sensitisation to enolases from distantly related organisms. These findings will contribute to an improvement of allergy diagnosis and patient-tailored prevention strategies.





