Abstract
Background: Eosiniphilic oesophagitis (EoE) is an immune-mediated disease with a complex pathophysiology. The accepted standard for objectively monitoring inflammation associated with this disorder is the number of eosinophils in oesophageal tissue biopsies obtained endoscopically. There is a need for alternative biomarkers that effectively correlate with disease activity and can hopefully be obtained non-invasively. The aim of this study is to review the literature on various biomarkers of EoE, with respect to their correlation to disease activity and response to treatment.
Methods: A literature search was performed using PubMed and OVID with keyword combinations of EoE and various potential biomarkers. Between 2006 and 2015, 39 studies that investigated the correlation of various tissue and serum biomarkers with EoE disease were identified.
Results: A number of candidates have emerged as potential biomarkers of inflammation in EoE. Eotaxin-3, interleukin (IL)-5, IL-13, microRNAs, and mast cell mediators have shown the most promise. Studies on these markers are quite heterogeneous in terms of methodology, with use of invasively as well as non-invasively obtained specimens.
Conclusion: The quest for an ideal biomarker for EoE continues. Establishment of normal values, effects of concomitant atopic diseases, age and gender, and validation of methodology of the tests are some of the challenges that future research should address.
INTRODUCTION
Eosinophilic oesophagitis (EoE) is a chronic immune-mediated disease characterised by oesophageal dysfunction, marked oesophageal eosinophilic infiltration (≥15 eosinophils/high-power field [hpf]), and variable response to acid suppression therapy.1,2 It affects both children and adults. The presenting symptoms, which vary by age, include difficulty eating, failure to thrive, chest and/or abdominal pain, dysphagia, and food impaction.3 Diagnostic guidelines published in 2007,1 and revised in 2011,4 require demonstration of oesophageal eosinophilia in the absence of gastro-oesophageal reflux disease (GORD) as determined through a clinical trial with proton pump inhibitors (PPIs) or the use of pH-monitoring studies. EoE is associated with other atopic diseases, and many patients have evidence of food and aeroallergen sensitivities. T helper 2 (Th2) cells and their associated cytokines are involved in the pathogenesis of EoE.3
Currently, oesophagogastroduodenoscopy and histological examination of oesophageal mucosal biopsies are required to establish the diagnosis, objectively assess response to therapy, document disease remission, and evaluate symptom recurrence.1,5 Reliable, non-invasive biomarkers have not yet been identified. An ideal biomarker for EoE should correlate with disease activity, reflect changes with therapy, have high sensitivity and specificity, be reproducible and cost-effective, and be performed on non-invasively and easily obtained specimen.5 A non-invasive biomarker continues to be elusive, although some promising candidates have emerged since our previous publication in 2012.6 In this article, we attempt to provide a summary of what is currently known about potential biomarkers of oesophageal inflammation among EoE patients. The complex pathogenesis of EoE provides numerous candidates. We have reviewed studies that used tissue and/or serum samples from patients who were diagnosed with EoE based on the widely accepted and practiced criteria per consensus recommendations published in 20071 and revised in 2011.4 A brief synopsis of the literature search is provided in Table 1. These studies not only have future diagnostic ramifications; they also provide valuable insight into the pathogenesis of EoE.
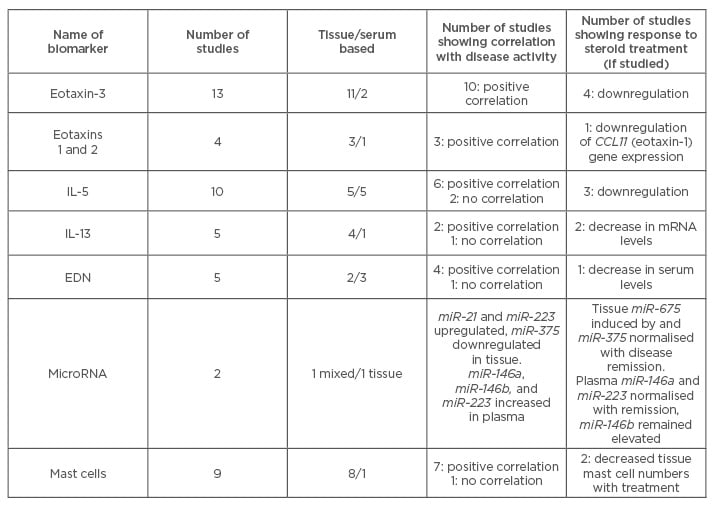
Table 1: Summary of studies on potential biomarkers for eosinophilic oesophagitis.
EDN: eosinophil-derived neurotoxin; IL: interleukin.
EOTAXINS
Eotaxins are chemoattractants shown to co-operate with interleukin (IL)-5 in tissue recruitment of eosinophils.7 Eotaxins 1, 2, and 3 have been investigated as potential biomarkers. The blood levels of eotaxins 1 and 2 did not correlate with oesophageal eosinophilia in a cross-sectional analysis of 47 paediatric patients with EoE.8 Studies on tissue expression of these two eotaxins have yielded mixed results in terms of correlation with disease activity.9-11 Eotaxin-3 however, has emerged as one of the most promising candidates as a biomarker for EoE. Oesophageal inflammation in EoE is driven by Th2 pathway in response to allergen exposure, leading to release of eotaxin-3 from oesophageal eosinophils.12 Genome-wide microarray expression analysis in 200613 showed markedly increased expression of eotaxin-3 encoding gene in EoE patients. Moreover, eotaxin-3 tissue expression was shown to positively correlate with oesophageal eosinophil numbers.14 IL-13 stimulation of oesophageal epithelial cells led to increased production of eotaxin-3.15 A study in 2008 demonstrated that eotaxin-3 oesophageal gene expression levels in EoE patients were down regulated upon steroid treatment.11 Eotaxin-3 messenger RNA (mRNA) levels had 89% sensitivity for distinguishing patients with and without EoE.16 Non-invasive analysis of blood eotaxin-3 levels showed significant correlation with oesophageal eosinophil density in a prospective cross-sectional analysis on 47 paediatric patients.8 Eotaxin-3 has been investigated as a marker to differentiate EoE from GORD. Bhattacharya et al.10 showed that the mean mRNA expression levels of eotaxin-3 were markedly elevated in patients with EoE as compared with patients with GORD and healthy control groups.
In a case control study, Dellon et al.17 used immunohistochemistry to compare the density of eotaxin-3 among EoE and GORD patients. Eotaxin-3 density was higher in EoE than in GORD, although the correlation with eosinophil count was weak. Another study published in 201418 investigated use of eotaxin-3 to differentiate EoE from proton pump inhibitor-responsive oesophageal eosinophilia (PPI-ROE). While oesophageal tissue from EoE patients showed significantly higher expression of eotaxin-3 compared with controls, it could not distinguish EoE from PPI-ROE. More recently, Moawad et al.19 also published data on a retrospective study on patients with EoE, GORD, and PPI-ROE. Eotaxin-3 staining scores were significantly higher for EoE patients compared to GORD (p=0.002), whereas there was a trend towards significance between EoE and PPI-ROE (p=0.054). Molina-Infante et al.20 studied gene expression of eotaxin-3 at baseline and after omeprazole 40 mg twice a day (b.i.d) for 8 weeks. Eotaxin-3 expression was indistinguishable between EoE and PPI-ROE at baseline. PPI therapy significantly decreased eotaxin-3 expression in PPI-ROE and in steroid-responsive EoE.20 Eotaxin-3 has been investigated for its potential as a non-invasive marker. There were no significant differences between serum levels of eotaxin-3 among EoE cases and controls, and among cases before and after treatment.21 There is a preponderance of evidence for the positive correlation of eotaxin-3 tissue expression and disease activity in terms of oesophageal eosinophilia and response to corticosteroid therapy, however, its potential as a non-invasive marker is less promising.
CYTOKINES
A number of studies have examined potential correlation of EoE with pathogenically related cytokines including IL-5, IL-13, IL-15, CCL5, and GM-CSF in the peripheral circulation.22 IL-5 plays a critical role in eosinophil trafficking into inflammatory sites along with other Th2 cytokines, IL-4, and IL-13.23 Tissue IL-5 mRNA expression levels were found to be significantly elevated in patients with EoE compared with GORD and controls.10 Lucendo et al.11 in 2008 showed that oesophageal IL-5 gene expression levels were variably downregulated after topical steroid treatment. Another study in 201116 showed a 4-fold induction of oesophageal transcripts of IL-5 receptor alpha in individuals with EoE. Serum levels of IL-5 in EoE patients have also been studied. In 2006, Konikoff et al.8 showed that blood levels of IL-5 did not correlate with oesophageal eosinophil density and were not increased in active EoE versus controls or those with inactive EoE. Bullock et al.14 showed that EoE patients with active disease had an increased percentage of CD4+ T cells expressing IL-5 compared with those in disease remission. Huang et al.24 demonstrated significantly increased levels of plasma IL-5 in EoE versus GORD. However, a longitudinal study in children with EoE showed that serum IL-5 levels in patients with EoE were not statistically different from those of the controls.25 IL-13 is a Th2 cell derived cytokine also involved in eosinophil trafficking to inflammatory sites.23 Gupta et al.9 in 2006 showed that IL-13 mRNA was similar between controls and patients with EoE. However, Blanchard et al.15 in 2007 found IL-13 mRNA levels to be markedly increased in oesophageal biopsy specimens from EoE patients compared with those from healthy individuals, with reversal following glucocorticoid treatment. In fact, the efficacy of anti-IL-13 therapy could be assessed with an IL-13-induced transcriptome. Moreover, IL-13 stimulation of oesophageal epithelial cells was shown to induce eotaxin-3 production.15 IL-5 and IL-13 gene expression in the oesophagus was studied at baseline and at 8 weeks following treatment with omeprazole 40 mg b.i.d in adult patients with EoE phenotype.20 PPI therapy significantly downregulated oesophageal IL-5 and IL-13 gene expression in PPI-ROE, similar to that seen in steroid-responsive EoE. Another study in 2011 identified IL-13 as one of the eight cytokines whose blood levels retrospectively distinguished EoE patients from healthy controls with 100% specificity and sensitivity. It proposed the development of a cytokine panel scoring system for predicting the diagnosis of EoE.16 Dellon et al.21 in 2015 evaluated serum IL-5, 6, 9, and 13 levels (among a panel of several serum biomarkers). No significant differences in assay values were seen between EoE cases and controls, or before and after treatment values among cases.21 Studies done so far on peripheral cytokine measurements do not establish a consistent correlation between peripheral levels and oesophageal disease activity. Moreover, peripheral cytokine measurement is affected by the confounding factors of concomitant allergic conditions; hence, it is very difficult to establish threshold levels. However, tissue expression of IL-5 and IL-13 mRNA could potentially be used for diagnosis as well as monitoring response to therapy in patients with EoE.
The cytokines could be useful targets for treatment options. The humanised monoclonal immunoglobulin (Ig)G antibody against human IL-5 (mepolizumab) has been investigated as a potential therapeutic intervention in EoE. In a randomised, placebo-controlled, double-blind trial in 2010, Straumann et al.26 demonstrated a marked decrease in mean oesophageal eosinophilia (p=0.03) in the mepolizumab group compared with placebo group 4 weeks after initiation of treatment which consisted of two intravenous infusions of 750 mg mepolizumab 1 week apart. Limited improvement of clinical symptoms was seen. Assa’ad et al.27 performed an international, multicentre, double-blind, randomised, prospective study of 59 children. Patients received an infusion every 4 weeks (a total of 3 infusions) of 0.55, 2.5, or 10 mg/kg mepolizumab. Peak and mean oesophageal intraepithelial eosinophil counts decreased significantly to 40.2±5.17 and 9.3±1.25 per hpf, respectively (p<0.0001). There was no placebo group in this trial.27 Rothenberg et al.28 investigated intravenous anti-IL-13 monoclonal antibody QAX576 in treatment of EoE. The mean oesophageal eosinophil count decreased by 60% with QAX576 versus an increase of 23% with placebo (p=0.004), and the decrease was sustained up to 6 months. There was a trend towards clinical improvement of dysphagia. While these results are encouraging, we need larger placebo-controlled trials to further define the potential therapeutic role of anti-IL-5 and anti-IL-13 antibodies in EoE.
PRODUCTS OF EOSINOPHIL DEGRANULATION
Eosinophil-derived neurotoxin (EDN) is an eosinophil granule-derived secretory protein with ribonuclease and antiviral activity.29 Yang et al.30 in 2008 demonstrated that EDN enhances antigen-specific Th2-biased immune responses. Extracellular EDN has been used as an indicator of eosinophil activation and degranulation in vitro. Increased levels of EDN in body fluids have been observed in patients with various eosinophil-associated diseases.31
A prospective, cross-sectional analysis on 47 paediatric patients undergoing endoscopic evaluation of possible EoE showed that plasma EDN levels significantly correlated with oesophageal eosinophil density and were increased in patients with active EoE versus controls.8 Similarly, Subbarao et al.25 found that serum EDN levels were significantly elevated in children with EoE compared with controls. However, in a recent study in 2015, serum levels of EDN were not found to be significantly different between EoE patients and control, and between pre and post-treatment EoE cases.21 Tissue EDN expression in EoE patients has also been examined. Alexander et al.32 in 2008 published a study on immunofluorescence (IF) staining of oesophageal biopsy specimens for EDN in four groups of patients: normal, low-level eosinophilia GORD patients, EoE patients with dysphagia with response to topical steroid therapy, and classic EoE patients with 25–100 eosinophils/hpf. EDN scores were found to be higher in the latter two categories, suggesting that extracellular EDN IF staining is a better marker for EoE than maximum eosinophil count.32 Another IF study suggested that tissue eosinophils may underestimate how extensively eosinophils are involved, particularly in individuals with marked eosinophil degranulation.31 The above studies do establish a positive correlation of tissue EDN with EoE disease activity, however further studies are needed to define its place in the diagnostic algorithm of EoE. Schlag et al.33 evaluated serum levels of eosinophil cationic protein (ECP) and mast cell (MC) tryptase as markers for response to topical steroid treatment in EoE patients. Serum ECP levels were shown to correlate significantly with oesophageal eosinophil counts compared with serum MC tryptase levels.
MICRO RNAS
MicroRNAs (miRNAs) are single-stranded RNA molecules 19–25 nucleotides long, that regulate post-transcriptional gene silencing of target genes.34 Multiple studies have demonstrated that EoE is associated with marked changes in tissue-specific gene expression, referred to as the EoE transcriptome.13,15 In 2012, Lu et al.35 showed that miRNA-21 and miRNA-223 were the most upregulated miRNAs in EoE patients, while miR-375 was the most downregulated. This miRNA signature correlated with the degree of tissue eosinophilia, was distinct from patients with chronic non-EoE, and was largely reversible with glucocorticoid therapy.35 Levels of miRNA-146a, miRNA-146b, and miRNA-223 were upregulated in plasma of EoE patients compared with healthy controls.35,36 The levels normalised with disease remission and inversely correlated with the degree of allergic inflammation.36 Interestingly, miRNA-375 expression inversely correlates with the degree of allergic inflammation in EoE, as measured by oesophageal eosinophil levels, the gene expression levels IL-5 and IL-13, and MC-specific enzymes.37 They further analysed oesophageal epithelial miRNA and mRNA from five paired biopsies pre and post-treatment with glucocorticoids, and found that 32 miRNAs were significantly upregulated and 4 were downregulated in pre-treated biopsies, with miRNA-214 being the most upregulated (150-fold).38 Taken together, these studies propose circulating miRNAs as promising candidates for non-invasive biomarkers of EoE due to their disease-specific dysregulation and their relative stability compared with mRNAs. Further studies will help refine their clinical utility.
MAST CELLS
MCs are important in the pathogenesis of EoE as they produce an abundance of cytokines that activate eosinophils and molecules that directly promote tissue remodelling.39,40 Studies quantifying intraepithelial MCs using anti-tryptase antibodies have shown higher counts among EoE patients than GORD and control groups.9,41 Abonia et al.42 generated transcriptome expression profiles of the MC proteases carboxypeptidase A3 and tryptase that were shown to correlate with MC levels and distinguished EoE patients from controls. Treatment of patients with fluticasone propionate normalised levels of MCs and MC transcriptome in responder patients.42 Aceves et al.43 showed significantly increased numbers of tryptase-positive MCs in oesophageal smooth muscle of EoE patients, with a significant reduction in numbers following the use of topical corticosteroid. Moreover, tryptase positive MCs were shown to express tumour necrosis factor (TNF)-β1 which increased contractility of cultured human oesophageal smooth muscle cells in vitro.43 In another study, expression of several MC-associated genes in biopsy specimens from patients with EoE without treatment were significantly increased compared with control subjects, followed by significant reduction by treatment with swallowed fluticasone.44 Dellon et al.17 in 2012 showed that patients with EoE had substantially higher levels of myelin basic protein (MBP) staining than GORD patients. Moreover, MBP density and eosinophil count correlated (r=0.81, p=0.001). In a subsequent study in 2014, oesophageal tissue from EoE patients was shown to have substantially higher levels of MBP and tryptase than controls on immunohistochemical analysis, compared with controls. However, these markers could not distinguish EoE from PPI-ROE.18 Dellon et al.21 in 2015 did not find any difference in plasma MBP levels between cases and controls, and between pre and post-treatment EoE cases. The above studies show that MC products and dysregulated transcriptomes in EoE patients could potentially be useful for disease diagnosis and monitoring.
ABSOLUTE EOSINOPHIL COUNT
Numerous studies have documented absolute and relative peripheral blood eosinophil counts in patients with EoE. The reported incidence of peripheral blood eosinophilia (PBE) shows a wide range of 10–100% over different age groups.5 Baxi et al.45 reported a 67% incidence of PBE in children and adolescents in 2006. A prospective cross-sectional analysis of paediatric patients undergoing endoscopic evaluation of possible EoE showed that absolute eosinophil count (AEC) levels significantly correlated with oesophageal eosinophil density.8 In general, the reported degree of PBE when present in patients with EoE is modest.5 While there appears to be a correlation between active oesophageal disease and peripheral eosinophil count, this parameter alone has limited potential as a disease marker. Especially since it may be affected by atopic disease in general, and may not be specific to oesophageal mucosal inflammation.22
There is very limited data correlating PBE with response to therapy. More data is needed regarding the effect of therapy for EoE on AEC in the context of eosinophil counts on oesophageal mucosal biopsies.
FRACTIONAL EXHALED NITRIC OXIDE
Fractional exhaled nitric oxide (FeNO) has been used clinically to monitor asthma inflammation. Results of FeNO were compared with oesophageal biopsy results among 51 patients. Exhaled nitric oxide was shown to have high specificity (87%) and negative predictive value (78%).46 In a prospective multicentre study, FeNO levels and symptom scores were measured among non-asthmatic EoE patients undergoing topical corticosteroid therapy.47 A statistically significant difference was found between pre and post-treatment FeNO levels (20.3 ppb [parts per billion] versus 17.6 ppb, p=0.009). However, the FeNO levels were not found to confidently predict a clinical or histological response.
OTHER MARKERS
Various other cytokines, chemokines, and cell products are under investigation as potential markers of disease activity and response to therapy. Protheroe et al.48 developed a monoclonal antibody specific to an eosinophil secondary granule protein eosinophil peroxidase and used it to identify intact eosinophils and detect eosinophil degranulation in formalin-fixed specimens. They developed a histopathologic scoring system to identify patients whose clinical course was suggestive of a diagnosis of EoE, but failed to reach the critical threshold of ≥15 eosinophils per hpf.48 Huang et al.24 studied plasma fibroblast growth factor basic levels by cytometric bead array to differentiate EoE from GORD. Another protein of interest is FK506-binding protein 51 (FKBP51). A study in 2010 demonstrated increased oesophageal FKBP51 mRNA levels in topical glucocorticoid responders compared with control subjects and patients with untreated active EoE, suggesting that increased FKBP51 transcript levels could be used to distinguish glucocorticoid responders from untreated patients with active EoE.49 Merves et al.50 reported increased expression of autophagy-related gene product 7 (ATG7) in active EoE compared with controls and EoE in remission. Zukerberg et al.51 demonstrated that intrasquamous IgG4 deposits may be useful as an adjunctive marker to distinguish GORD from EoE. Intrasquamous extracellular IgG4 deposits were seen in 76% of EoE cases compared with none of the GORD cases. Moreover, EoE patients on treatment were less likely to be positive for this marker.51 Group 2 innate lymphoid cells are a recently discovered group of lineage-negative cells that express the chemoattractant receptor homologous molecule expressed on Th2 lymphocytes. Recently it has been shown that these cells can be successfully quantified in oesophageal biopsy specimens, and their expression is significantly higher among patients with active EoE versus inactive EoE, and also higher than expression among controls and PPI-ROE patients.52 The oesophageal string test was investigated as a minimally invasive clinical device to measure oesophageal inflammation among children by Furuta et al.53 MBP-1, EDN, ECP, and eosinophil peroxidase were measured in luminal effluents eluted from oesophageal string tests and extracts of mucosal biopsies. The levels of eosinophil-derived proteins in luminal secretions were found to be reflective of mucosal inflammation. The immune mechanism involved in the pathogenesis of EoE is associated with differential expression of various inflammatory and epithelial-derived genes. Matoso et al.54 used gene expression microarray and reverse transcription polymerase chain reaction to screen oesophageal biopsies from paediatric EoE patients before and after treatment with topical steroids. Overexpression of ALOX15 and TNF-a-induced factor 6, and underexpression of filaggrin, SLURP1, and CRISP3 was noted in EoE. They subsequently demonstrated that positive ALOX15 expression is more prevalent in EoE than in GORD, and could be a valuable marker to differentiate between the two conditions.
CONCLUSIONS
The current standard of care for diagnosis of EoE and monitoring of response to treatment dictates the enumeration of eosinophils in a mucosal biopsy specimen obtained endoscopically. The pathogenesis of EoE is complex and hence a plethora of biomarkers are being studied to objectively assess the inflammation associated with this disease. The above review attempts to summarise the vast body of literature available on potential biomarkers for EoE. Eotaxin-3, IL-5, IL-13, and lately, miRNAs have all shown promise in this direction. The search continues for a biomarker which could be obtained by a non-invasive or minimally invasive means, and accurately reflect the clinicopathologic diagnosis of EoE. Establishment of normal values, effects of concomitant atopic diseases, age and gender, and validation of methodology of the tests, are some of the challenges that future research should address.






