BACKGROUND
Allergic diseases are becoming a public health concern, their frequency has notably increased in the last decades, which is likely related to changes in lifestyle and environment. In recent years, the epigenetic regulation has emerged as a pivotal group of mechanisms that can help explain the allergic disease emergence and allows us to understand their molecular basis.1
Some of these mechanisms of epigenetic regulation are conducted by small non-coding RNAs (sncRNA). Small cytoplasmic RNA, or Y-RNA, are a group of sncRNA of approximately 100 bp in length and are highly conserved from the evolutionary point of view and involved in the initiation of DNA replication and RNA stability that regulate gene expression.2,3 Besides their cellular functions, their presence in the extracellular milieu, as part of ribonucleoprotein complexes or associated to extracellular vesicles, underscores their potential role in the modulation or amplification of different responses whether at local or at systemic level.4 Thus, Y-RNA have
been previously related to autoimmunity5 and cancer,6 and now to allergy. In this sense, our group has recently found differential Y-RNA expression profiles in allergic patients. In particular, the authors have shown a significant increase of certain Y-RNA in pollen allergic patients.7
The proper study of epigenetic regulation
requires the implementation of the new methods for its research. To increase the repertoire of laboratory approaches to the disease, cell line studies need to be improved with new sophisticated techniques that mimic pathological states to deepen their molecular mechanisms. To unravel the function of these small RNA in allergy, we have developed a strategy for transiently silencing Y-RNA in cell culture. This will allow us to study the physiological effects behind different expression levels of the
Y-RNA of interest in this particular cell line, as well as how it is related to signal transduction mediated by extracellular media.
METHOD
Transient silencing of different Y-RNAs was performed in the cell line Jurkat, used as a model of T lymphocyte. Dicer-substrate short interfering RNA (DsiRNA) were used; these are 27mer
duplex RNA that demonstrate increased potency in RNA interference compared to traditional,
21mer short interfering RNA8 (provided by Integrated DNA Technologies, Inc., USA) designed against the Y-RNA of interest.
Cells were plated at a density of 2.5 x105 cells/mL and transfected with DsiRNA designed for each Y-RNA or the scrambled control at a final concentration of 50 nmol/L. Briefly, DsiRNA were diluted in of Opti-MEM® (Thermo Fisher Scientific, USA) and mixed with Lipofectamine® RNAiMAX (Thermo Fisher Scientific, USA) diluted in Opti-MEM®. The complexes of DsiRNA-Lipofectamine® RNAiMAX were added directly to the cells and mixed gently. Complexes did not have to be removed following transfection. Cells were incubated at 37 °C in a CO2 incubator for 3 days post transfection before assaying for silencing gene expression.
RESULTS
With this method successful and significant silencing of the different Y-RNA was obtained. When compared to the scrambled transfected control, the levels of the specific transcript were reduced between 32% and 85% (Figure 1).
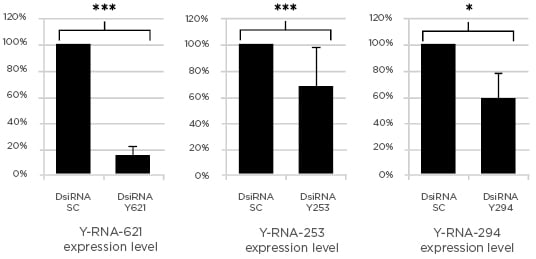
Figure 1: Changes in Y-RNA levels after transfection with Dicer-substrate short interfering RNA.
DsiRNA: Dicer-substrate short interfering RNA (***p<0.001; *p<0.05).
CONCLUSION
These findings have led to the development of an easy and affordable method to silence sncRNA in the cell line Jurkat, providing new perspectives in this cell line to study molecular mechanisms related to allergic diseases and its epigenetic in vitro control.





