BACKGROUND AND PURPOSE
In critically ill patients, colonisation by multidrug-resistant (MDR) bacteria is a significant risk factor for the development of subsequent infections.1-3 Active surveillance cultures are routinely used to screen for MDR bacteria in selected scenarios.4 The purpose of this study was to evaluate the epidemiological and molecular factors common between colonising bacteria and healthcare-associated infection (HAI) events caused by the same MDR bacteria in critically ill patients.
METHODS
The study was performed between January 2016 and May 2018 in two intensive care units (ICU), clinical and surgical, of a public tertiary hospital which specialises in cardiologic care. All patients admitted to these ICU had active surveillance cultures collected on entrance and then weekly, for the practice of contact precautions in cases of MDR bacteria detection.
The bacterial identification and susceptibility profiles were determined using Vitek® MS and
Vitek® 2 Systems (bioMérieux, Marcy-l’Étoile, France), respectively; the detection of carbapenemases, vanA, and vanB genes were determined by real-time PCR. If a patient developed a HAI, comparison between antimicrobial susceptibility profiles of colonising and infecting strains was made. If available, genetic relatedness of the strains was characterised by pulsed-field gel electrophoresis. The HAI were characterised according to Centers for Disease Control and Prevention (CDC) criteria.
RESULTS
Thirty-six patients colonised by MDR bacteria who developed HAI by MDR were included in the study. Twenty-eight HAI episodes were caused by the same colonising bacteria with an identical antimicrobial susceptibility profile. The other eight HAI episodes were caused by non-concordant MDR microorganisms. The mean time between MDR colonisation and infection was 21.9 days. Carbapenemase-producing Klebsiella pneumoniae was the most frequently detected bacterium (n=24; 85.71%), followed by vancomycin-resistant Enterococcus spp. (n=2; 7.14%), carbapenem-resistant Acinetobacter baumannii (n=1; 3.57%), and Pseudomonas aeruginosa (n=1; 3.57%). The most prevalent HAI were central line-associated bloodstream infections in patients in the clinical ICU, and surgical site infections in patients in the surgical ICU. Fourteen episodes could be analysed by pulsed-field gel electrophoresis and the concordance rate was 100%. The colonising pathogens and HAI distribution are shown in Table 1.
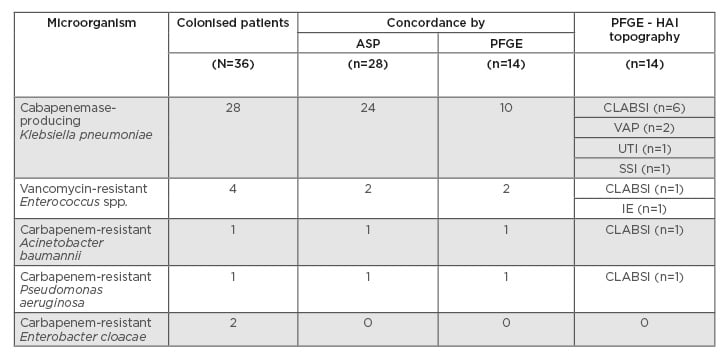
Table 1: Colonising and infective bacteria concordance.
ASP: antimicrobial susceptibility profile; CLABSI: central line-associated blood stream infection; HAI: healthcare-associated infection; IE: infective endocarditis; PFGE: pulsed-field gel electrophoresis; SSI: surgical site infection; UTI: urinary tract infection; VAP: ventilator-associated pneumonia.
CONCLUSION
MDR colonisation prevention measures are essential and must be performed in critically ill patients. Colonisation by MDR bacteria can be predictive of subsequent infection aetiology. An improvement in empiric antimicrobial treatment adequacy can be achieved if active surveillance culture is performed routinely.








