Abstract
Type I IFN have important roles in many paediatric and adult rheumatic diseases and are a new therapeutic target for which several anti-IFN treatments are currently in use or in development. Since the direct detection of these proteins in biological samples has proved challenging, indirect methods are often used to infer the presence of type I IFN. Most commonly this involves quantification of the relative expression of interferon-stimulated genes (ISG) that are used to calculate an interferon score (IS).1 This score has been used for example to asses type I IFN activity in paediatric patients with type I interferonopathies, systemic lupus erythematosus, dermatomyositis, and systemic juvenile idiopathic arthritis.2 Both quantitative PCR (qPCR) and NanoString technology have similar sensitivity and reproducibility for IS determination.3 The use of different whole blood RNA collection systems on the IS has not been evaluated despite evidence of method-dependent changes in gene expression.4
The aim of the study presented at the European League Against Rheumatism (EULAR) 2019 congress in Madrid, Spain, was to compare expression of six common ISG (IFI27, IFI44L, IFIT1, ISIG15, RSAD2, and SIGLEC1) and the corresponding IS in RNA derived from two commonly used whole blood RNA collection systems (PAXgene [PreAnalytiX, Becton Dickinson] and Tempus [Applied Biosystems]).
For the purpose of the study, whole blood was collected from 10 healthy individuals (median age 25.5 years) in sodium heparin tubes and incubated with or without recombinant human IFN alpha 2b (rhIFNα, 2 IU/mL, 4 hours, 37 °C, 5% CO2). Next, samples were divided between PAXgene and Tempus tubes and RNA was isolated according to the manufacturer’s protocols. cDNA was synthesised (~500ng input RNA; qScript cDNA synthesis kit) and ISG expression measured on a QuantStudio 6 Real-Time PCR instrument using a TaqMan Fast Advanced Assay. For each ISG, expression was normalised against the geometric mean of two housekeeping genes (18s rRNA and HPRT1) and calculated using the formula 2-∆Ct. Relative gene expression was reported as the normalised expression of each ISG divided by the median of normalised expression of the same ISG in unstimulated samples. The median relative expression of all six ISG was used to calculate the IFN score for each sample.
The results showed that there was no statistically significant difference in the expression of any of the six ISG in either the rhIFNα-stimulated or unstimulated samples derived from PAXgene or Tempus tubes. Overall there was a strong correlation of the IFN score between PAXgene and Tempus tubes for both the unstimulated (R2=0.9117, p<0.0001) and rhIFNα-stimulated samples (R2= 0.8529, p=0.0001).
Despite reported differences in gene expression patterns associated with samples collected in PAXgene versus Tempus tubes, the results demonstrated that 6-gene IFN scores do not differ significantly between RNA samples obtained with these two systems. These results suggest that health care and research centres can use either tubes for IFN score determination using these 6 ISG and results can be directly compared irrelevant of the RNA collection system employed.
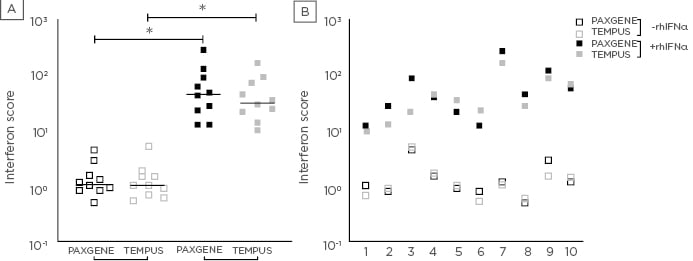
Figure 1: Interferon score derived from PAXgene and Tempus tubes.
Interferon score (y-axis) calculated for 10 healthy individuals (x-axis; A) following 4-hour ex vivo incubation of whole blood in the absence (open squares) and presence (solid squares) of rhIFNα and subsequent collection in PAXgene (black) and Tempus (grey) tubes (x-axis; B). Horizontal lines represent the median interferon score (n= 10; B). *indicates p <0.005.