Abstract
Introduction: Hand hygiene is one of the most effective and inexpensive means of preventing the spread of communicable diseases. Rates of handwashing worldwide are low, and poor handwashing practices in universities remain a public health challenge.
Objective: The objective of this study was to examine the practice of handwashing, the microbial communities, and the susceptibility pattern of micro-organisms isolated from the palms of students of the Obafemi Awolowo University in Ile-Ife, Osun, Nigeria.
Methods: A combination of qualitative and quantitative methods was used to retrieve data. A self-administered questionnaire was utilised to gather socio-demographic characteristics and the practice scale of handwashing from the respondents. Isolation and identification were carried out by culture-based surveys and biochemical tests. Disk diffusion was used to determine susceptibility.
Results: The majority of respondents were between ages 21–25 years, with 54.3% of them being female. About half of individuals never used an alcohol-based hand sanitiser, while only 4.9% of respondents admitted to always using soap to wash their hands. Presumptive identification of the organisms showed 38.1% of organisms as Staphylococcus epidermidis and 17.58% as Micrococcus spp. None of the antibiotics showed total efficacy. The resistance to chloramphenicol was low, and <50% of the isolates showed resistance to tetracycline, novobiocin, and sulphonamide. Resistance to nalidixic acid was seen in 58.4% of organisms tested, and 77.75% were susceptible to tetracycline.
Conclusion: Instances of improper hand hygiene were high, and this may increase the spread of micro-organisms through hand carriage. High resistance to the antibiotics tested was prevalent. Local trends of antimicrobial resistance must be robustly studied and proper interventions developed.
Key Points
1. Misuse of antibiotics has led to increasing antibiotic resistance. A significant strategy to reduce the misuse of antibiotics, and thus reduce antibiotic resistance, is to decrease the transmission of micro-organisms via hand carriage. Therefore, it is important to assess handwashing practices of individuals, as well as the microbial spectrum and susceptibility of micro-organisms isolated from the palmar surface.
2. This study was a cross-sectional study targeting the undergraduate students of a tertiary institution in Nigeria to assess their handwashing habits, and to determine the susceptibility of isolated palmar micro-organisms to selected antibiotics.
3. Results indicated poor hand hygiene practices and high resistance to the antibiotics tested. Local patterns of antibiotic resistance must be studied, and programmes for antibiotic stewardship put in place to deal with the increasing prevalence of antimicrobial resistance.
INTRODUCTION
Resistance of bacteria to antibiotics is an issue that has plagued clinicians since the discovery of antibiotics. Antibiotic resistance is the ability of bacteria to resist the toxic effects of drugs, and grow in the presence of a concentration that will normally kill or inhibit its growth.1 This leads to higher medical costs, prolonged hospital stays, and increased mortality. The World Health Organization (WHO) cautions: “Without urgent action, we are heading for a pre-antibiotic era, in which common infections and minor injuries can once again kill.”2
In the golden age of antibiotics, this problem was easily solved by the discovery, or synthesis, of new antibiotics. In recent years, however, the production of novel antibiotics has slowed down immensely, and the introduction of novel therapeutic agents is outpaced by the ever-evolving microbes.3 The development of new antibiotics takes years and millions of dollars, only for the drug to be active for a relatively short period of time, and its activity reduced or eliminated by microbial resistance. Pharmaceutical companies have thereby diverted funds into more rewarding areas such as the development of drugs for chronic illnesses.4
Antibiotic stewardship is an important part of controlling the improper use of antibiotics and thus, extending their lifespan. The WHO suggests the establishment of antimicrobial stewardship committees in healthcare facilities, and the dedicated collection of data to “assess the extent and quality of antibiotic use, identify problematic prescribing practices, and compare appropriate use […] over time.”5
Another significant strategy in reducing the misuse of antibiotics is reducing the rate of pathogen transmission via hand carriage. Pathogen transmission is broken down into five steps: 1) organisms must be present on the skin or inanimate objects in the immediate vicinity; 2) organisms must be transferred to the hands; 3) organisms must be able to survive on the carrier’s hands; 4) failure to carry out proper hand antisepsis; and 5) the carrier’s contaminated hand deposits organisms on another individual or inanimate object that will come in contact with another individual. Hand hygiene, when practised at any of these stages, will drastically reduce the hand carriage of micro-organisms.6
Proper hand hygiene involves the use of an alcohol-based sanitiser containing at least 60% alcohol for 20–30 seconds, or the washing of hands with soap and water for a minimum of 40 seconds. To ensure the hands have been thoroughly cleansed when washing, the Africa Centres for Disease Control and Prevention (Africa CDC)7 suggests wetting the entire surface of the hands with warm or cold running water, lathering the backs of the hands, between the fingers and under the nails, with soap, rubbing the hands together for at least 20 seconds, and rinsing completely with clean, running water.
Several studies have been conducted on handwashing knowledge, attitudes, and practices of individuals. It was shown by Duong et al.8 that, while most participants knew the usefulness of handwashing in curbing the spread of infection, less than half regularly washed their hands daily. This study further noted that community education would emphasise proper handwashing behaviour.
It is not in all situations that awareness of the usefulness of handwashing was found to be sufficient. A study conducted in Saudi Arabia showed that only 46% of students were of the opinion that handwashing could protect against disease, and 34% of the correspondents in that study thought handwashing was only useful for removing dirt.9 The study also showed that many students received handwashing information from their parents.
In the results published by Sultana et al.,10 it was shown that the practice of handwashing could be improved by the removal of obstacles to hand hygiene, such as low availability of soap and adequate water supply within the university.
Due to the major role the hand carriage of micro-organisms plays in the dissemination and selection of resistant micro-organisms, the hand washing awareness of the public must be assessed. If the degree of awareness is high, the spread of micro-organisms reduces, and vice versa. Once the degree of hand washing awareness has been assessed, then crucial steps towards the improvement of public health knowledge and sanitation can be taken.
The WHO states that the “failure to perform appropriate hand hygiene is considered the leading cause of hospital-acquired infections (HAI) and spread of multidrug-resistant organisms, and has been recognised as a significant contributor to outbreaks.”6 Therefore, it is important that awareness of the usefulness of hand washing by the populace be evaluated and the microbial community present on the hands identified and characterised. This study assesses the hand washing practices of a selected student population of the Obafemi Awolowo University, Ile-Ife, Osun, Nigeria, and determines the microbial spectrum as well as the susceptibility of the micro-organisms isolated from the palmar surface of those students.
MATERIALS AND METHODS
Study Location
This study was conducted at the Obafemi Awolowo University (OAU), Ile-Ife, Osun State, Nigeria. The University is one of the largest tertiary institutions in Nigeria, with about 35,000 students.
Study Design
This was a cross-sectional study of male and female students between the ages of 18–40 years at Obafemi Awolowo University. The study comprised both the filling of questionnaires and laboratory experiments. The questionnaire was adapted and developed from a previous study,11 and included close-ended questions on the attitude and practices of students when washing their hands.
Study Population
The sample size was calculated using Cochran’s population proportion formula with the following assumptions: proportion of individuals who have transient bacterium present on their palms (p)=95% from a similar study;12 95% confidence interval; z=the standard normal tabulated value; and desired level of precision (margin of error; d)=5%.
Cochran’s formula:
N= z2p(1-p)
![]()
d2
The calculated sample size was approximately 72 participants. To make allowance for non-response, a total number of 81 pretested closed-ended questionnaires were administered.
The criteria for inclusion were participants within the specified age of 18–40 years, participants in classes 100–500 levels, and participants registered full-time in the university. Students were excluded if they did not meet the inclusion criteria, had physical disabilities, had visual and hearing impairments, and were post-graduate students.
Data Collection
The students were approached, and the purpose of the study, as well as what was required of them, were explained thoroughly. Students who volunteered for the study were introduced to the questionnaire, its purpose, and how to fill it. Confidentiality was assured to enable students to fill out the questionnaire accurately and without bias. The questionnaire was self-administered by the students.
Sample Collection and Processing
The various samples were collected over a period of 14 days (15th–28th June 2021) at various sites in the Obafemi Awolowo University.
The palmar surfaces of both hands of the students were swabbed with a sterile swab stick moistened with a solution of sterile 0.1% Tween™ 80 (Croda International, Snaith, UK). The entire palm surface was swabbed in two perpendicular directions to ensure that the maximum surface area of each palm was represented in the sample. The swabs were then suspended in 2.5 mL of the same solution in universal bottles, and shaken on a wrist action shaker for 5 minutes.13
The samples were streaked onto the surface of plates of four different agars: nutrient agar, which serves as a general-purpose growth medium for the isolation and cultivation of bacteria; mannitol salt agar, which selectively allows for the growth of gram-positive bacteria such as Staphylococcus and Micrococcaceae; eosin-methylene blue agar, which serves as a growth medium for the selective growth of gram-negative bacteria; and MacConkey agar, which allows for the selective growth of gram-negative bacteria and differentiates the bacteria based on their ability to ferment lactose. These plates were incubated for 48 hours at 37 ºC. The grown colonies from the nutrient agar plates were then sub-cultured on other freshly prepared nutrient agar plates and incubated at 37 ºC for 48 hours. The various morphological characteristics of the colonies such as size, shape, colour, elevation, surface, and margin were noted.14 The isolated colonies were then stored on nutrient agar slants and cryopreservative medium for further use.
The isolates were gram-stained and subjected to several biochemical tests (catalase, coagulase, indole production, blood agar, biofilm formation, and triple sugar iron tests) to identify them.15
Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was carried out on all isolates and the disk diffusion method was employed (Kirby–Bauer technique) with Muller–Hinton Agar (Oxoid Ltd, Hampshire, UK).16 The antibiotics tested include tetracycline (30 µg), chloramphenicol (30 µg), novobiocin (5 µg) for 247 of the isolates; and nalidixic acid (30 µg), trimethoprim (2.5 µg), compound sulphonamide (300 µg) for 113 of the isolates. The results were obtained by measuring the zone of inhibition and comparing it with the Clinical and Laboratory Standards Institute (CLSI) 2016 interpretative performance standard for antimicrobial disk susceptibility testing.17,18
Only six antibiotics were used in the susceptibility testing, due to the financial limits of the project. This study was conducted after trade in Nigeria commenced after the COVID-19 pandemic and there was a shortage of antibiotic discs. Due to this, cephalosporins, carbapenems, vancomycin, metronidazole, and combinatorial antibiotics were not available for purchase.
Data Analysis
Data collation was done using Microsoft Excel 365 (Microsoft, Redmond, Washington, USA). Analysis was carried out using SPSS Version 21 (IBM, Armonk, New York, USA). The descriptive statistics used include the mean, standard deviation, frequency, and mean weighted averages. Bar charts were used where necessary.
RESULTS
Sociodemographic Characteristics of Participants
All 81 questionnaires administered were retrieved. Table 1 provides details on the socio-demographic characteristics of the study population. On the sex of the respondents, 54.3% of the respondents were female. The age of the respondents ranged from 16–34 years, with a mean of 21.2±0.9 years.

Table 1: Attitude to hygiene and hand washing practices.
Attitude to Hand Hygiene and Hand Washing Practices
Concerning their attitude to hand hygiene, the majority of students (34.6%) washed their hands frequently, and 22.2% of the students washed their hands twice a day. Table 2 shows that a slight majority of respondents washed their hands for about 20 seconds to 1 minute, while 40.7% of the respondents washed their hands for less than 20 seconds. The calculated average length of time spent hand washing was 27.8±2.4 seconds.

Table 2: Antibacterial susceptibility pattern of palmar isolates.
As shown in Table 1, when questioned about how long ago they practised hand hygiene, 82.7% responded that they last washed their hands over half an hour ago, 9.9% responded less than 15 minutes ago, 4.9% of the respondents responded 5–15 minutes ago, and 2.5% of the respondents responded 15–30 minutes ago. Many of the respondents (39.5%) sometimes washed their hands after using the toilet, and 39.5% of the respondents sometimes washed their hands after eating. Most of the respondents (66.7%) sometimes washed their hands after returning to their room or house after school, with a similar proportion of the respondents never using soap to wash their hands. Most of the respondents (54.3%) never used a hand sanitiser (alcohol-based) to cleanse their hands, and a vast majority (95.1%) of respondents never used hot water when washing their hands. About half of the students (51.9%) wiped their hands after washing, with 39.8% of the respondents drying by shaking the water droplets off their hands. A majority of respondents noted that the coronavirus outbreak affected their hand-washing practices, and acknowledged that their frequency of handwashing increased.
Distribution and Presumptive Identification of the Isolates
Regarding the number of micro-organisms isolated from the palmar surface of the study participants and the presumptive identification of micro-organisms isolated, Staphylococcus epidermidis was the predominant isolate, with 38.10% of the micro-organisms present on the respondents’ palms; Micrococcus spp was a distant second, with 17.58% of the isolates; a total of 11.72% were Staphylococcus aureus; 8.43% were Corynebacterium spp; 5.86% were Streptococcus spp; and 5.13% were from the Bacillus genus.
The organisms present in the smallest numbers were Listeria monocytogenes (2.56%), Neisseria spp (1.88%), Klebsiella/Enterobacter spp (1.47%), Haemophilus spp (1.10%), and Pseudomonas/Proteus spp (1.10%), with Staphylococcus saprophyticus and Escherichia coli being the rarest isolates (0.73%).
Figure 1 depicts the distribution of the micro-organisms isolated from the palmar surfaces of the study population.
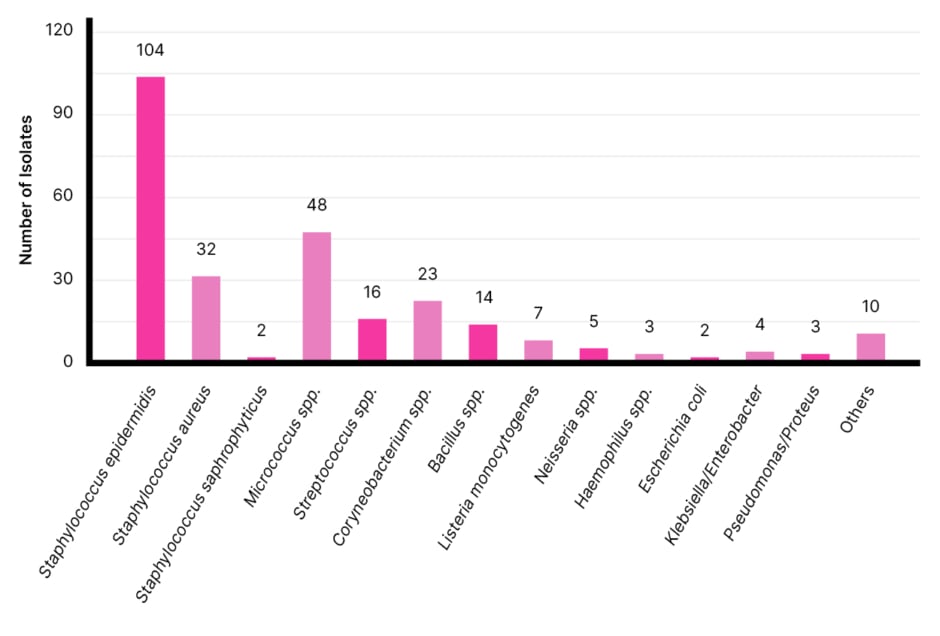
Figure 1: Presumptive identification of isolates.
Susceptibility and Resistance Pattern of Isolates
In Table 2, the general susceptibility and resistance pattern of the isolates are described. The isolates were most sensitive to chloramphenicol, regardless of whether they were gram-positive or gram-negative. For the gram-positive organisms, over 70% of the isolates were susceptible to tetracycline and compound sulphonamide. Nalidixic acid and trimethoprim showed the least efficacy, with only 39.8% of the gram-positive isolates being susceptible. The gram-negative isolates were the least susceptible to trimethoprim and compound sulphonamide (41.7%).
DISCUSSION
The objectives of this study were to describe the handwashing practices of students of Obafemi Awolowo University, to characterise the hand flora in terms of bacteria present, and to determine the patterns of resistance of the isolated micro-organisms to selected antibiotics.
Regarding the hand-washing practices of participants in the study, results indicate that most respondents maintained a form of hand hygiene, including washing hands with or without soap, but most respondents failed to meet the WHO standards for proper hand hygiene, with the majority washing their hands for the sufficient length of time, but without soap or warm water.9
It was also noteworthy that most individuals did not use an alcohol-based hand sanitiser. Many students also agreed to washing their hands rarely, once or twice a day, or whenever necessary. This is an insufficient number for proper hand hygiene and is not sufficient to prevent the spread of infection through hand carriage.19,20
On culturing in nutrient agar, a total of 273 colony-forming units (CFU) were isolated from the palmar surfaces of the 81 students, with the majority of individuals having more than 3 CFU on their hands. Out of a total of 273 CFU, a large majority (93.77%) were gram-positive, and only 6.23% were gram-negative. Staphylococcus spp. was the most abundant genus to be isolated from the students’ palmar surfaces, with S. epidermidis having the highest occurrence. Micrococcus spp. was the second most abundant organism isolated, followed by Corynebacterium spp., Streptococcus spp., Bacillus spp., and Listeria spp.
These results were in accordance with literature that states that S. epidermidis is one of the most abundant micro-organisms present on the skin.21 The identity of the other organisms isolated is also in line with previous studies on skin bacteria, although in different population numbers.22,23 This confirms that while the bacterial flora present on the skin is diverse, there are a few constant genera that are considered to be residential bacteria. Transient bacteria have been shown to often be a reflection of the environment.
It has, however, been observed that the skin genus Staphylococcus is more readily cultivated than organisms like Corynebacterium spp., and this difference in growth might lead to potential erroneous results from culture-based surveys. To overcome this shortcoming, sequencing methods should be henceforth applied to identify members of microbial communities.24
Many of the resident bacteria do not cause disease but some transient micro-organisms are found on the skin that have high pathogenicity and can cause diseases in immunosuppressed individuals.25 There is also a possibility that transient microbe exposure could result in tissue damage and allergic, inflammatory, and autoimmune responses, even after the causative organism is no longer present.26 An important example is S. aureus. The CDC estimated that about 33% of individuals carry S. aureus. S aureus can produce clinically important enzymes such as haemolysins and leucocidins that aid in its pathogenicity.27
Most of the bacteria that inhabit the skin are gram-positive, but there are a few gram-negative bacteria that are found, although infrequently, on the skin. An important example is Pseudomonas aeruginosa (which 1.10% of the isolates in this study were identified as), a gram-negative rod that is a known pathogen. In addition to being associated with leg ulcers, P. aeruginosa is one of the leading causes of morbidity and mortality in burn victims.28
The antimicrobial resistance and susceptibility pattern of the organisms isolated on the palmar surfaces is also crucial, as these organisms can cause serious infections upon entering the bloodstream. Listeriosis, a rare but severe foodborne disease, is caused by Listeria monocytogenes, another organism isolated in this study. Previous studies in the region have shown a high level of resistance of L. monocytogenes isolates to commonly used antibiotics.29 Palmar isolates can also aggravate wounds, infecting them, increasing the time it takes to heal, and reducing the effectiveness of antibacterial agents used. In general, gram-positive bacteria showed more sensitivity to the antibacterial agents tested.30
In the interpretation of the results, the number of organisms that displayed an intermediate reaction to the antibiotics will be grouped as resistant to give a clearer picture of the resistance/susceptibility pattern.
Tetracycline was selected for inclusion in the study because it is one of the most commonly used (and misused) antimicrobials in the country.31,32 Only 20.2% of gram-positive isolates showed resistance to tetracycline, but that proportion increased to 41.7% in the gram-negative isolates tested.
A study done by Mama et al.33 found that the majority of organisms isolated displayed resistance to nalidixic acid, while a minority showed resistance to chloramphenicol. This is comparable with the results obtained in this study. The resistance of the isolates to chloramphenicol was low in this study (12.96%). Although resistance to this antibiotic is well documented, the low figures of resistance obtained can be attributed to the fact that chloramphenicol is not commonly prescribed due to its side effects.34
Generally, less than 50% of the isolates showed resistance to tetracycline, novobiocin, and sulphonamide. The majority of the isolates tested displayed resistance to trimethoprim. No antibiotic was 100% effective against the isolates.
The high resistance to trimethoprim could be related to its bacteriostatic mode of action and the fact that it is a competitive inhibitor, whose effectiveness will reduce in higher concentrations of dihydrofolate (the substrate it inhibits). When combined with sulphonamides, it is expected that the resistance would reduce, since a bactericidal effect would result.35
Trimethoprim and sulphonamide combination drugs are one of the most prescribed antibiotics in low- and middle-income countries like Nigeria, and this leads to the high levels of resistance seen in this study and in similar studies conducted in the region.36,37
A significant proportion of the gram-positive cocci isolated from the study participants were coagulase-negative Staphylococci (CoNS). CoNS, including S. epidermidis, are often referred to as ‘accidental pathogens’. However, in recent years, CoNS have been reported to cause clinically significant ocular symptoms.38 Antibiotic resistance in CoNS is of utmost importance because CoNS, such as S. epidermidis, inhabit the same ecological niche as the pathogenic S. aureus, and may serve as reservoirs of genes that facilitate methicillin-resistant S. aureus (MRSA) infection after horizontal gene transfer.39
As indicated in the results, CoNS are more susceptible to antibiotic action, but susceptibility to nalidixic acid, trimethoprim, and compound sulphonamide is low for both species in the Staphylococci genus, while susceptibility to tetracycline is relatively high. This is in contrast to other studies, such as by Petrillo et al.,38 which showed high susceptibility to trimethoprim and sulphonamide, whilst recording high resistance to tetracycline. Differences in prescriber habits and over-the-counter use may be the reason for the variation in antibiotic resistance trends. Similar studies in Nigeria40 show moderate resistance to chloramphenicol at 45%.
Novobiocin was mainly used to differentiate between S. saprophyticus and other coagulase-negative Staphylococci. This test exploits the intrinsic resistance of S. saprophyticus to the antibiotic. The basis of this inherent resistance is, however, unknown.41 Catalase-positive and coagulase-negative Staphylococci that present with little or no zone of inhibition for novobiocin were presumptively identified as S. saprophyticus. Although simple, a limitation of this test is the fact that occasional human isolates that are not S. saprophyticus may also be resistant to novobiocin. It is therefore recommended that other biochemical, molecular, or immunological methods of testing be employed for confirmatory identification.42
CONCLUSION
The results obtained in this study showed the handwashing practices, spectrum, and susceptibility pattern of the microbial communities isolated from the palmar surfaces of students of the University. Results indicate poor hand hygiene practices, and these have the tendency to lead to the spread of micro-organisms through hand carriage. Among the isolates obtained from the hands of the respondents, Staphylococcus spp. was the most abundant genus, with S. epidermidis having the highest occurrence. High resistance to the antibiotics tested was shown with trimethoprim, nalidixic acid, and novobiocin showing the lowest efficacy. The isolates were most susceptible to chloramphenicol, tetracycline, and sulphonamide. Local patterns of antibiotic resistance must be studied, and programmes for antibiotic stewardship put in place to deal with the increasing prevalence of antimicrobial resistance.
STUDY LIMITATIONS
This study did not make use of genotypic testing to conclusively identify the organisms isolated. As such, the isolates could only be identified phenotypically. Additionally, the use of culture-based surveys might produce isolates that are more easily cultivated and not a true reflection of the palmar community.
ETHICAL APPROVAL
Ethical clearance was obtained from the Health Research Ethics Committee, Institute of Public Health, College of Health Sciences, Obafemi Awolowo University, Ile-Ife, Osun state, with the certificate number being IPH/OAU/12/1736.







