Abstract
Emerging technologies are profoundly changing the landscape of allergy diagnoses and future allergy treatments. At the single patient level, the introduction of single components and allergen microarrays for allergy diagnoses has significantly modified treatment strategies. In epidemiological terms, the availability of information from large dataset analyses has allowed and, more importantly, will allow for changes in prophylaxis and treatment strategies in many patients. In this report, we describe the different fields where new technologies have had a significant effect on allergy management and identify new scenarios where the combination of data from basic, clinical, and epidemiological research will improve our knowledge of allergy diagnosis and treatment.
INTRODUCTION
Allergen immunotherapy (AIT) was empirically described more than 100 years ago and immediately showed its clinical efficacy in controlling allergic respiratory diseases.1,2 Its efficacy is, however, strictly related to a correct aetiological diagnosis. Fifteen years ago, the introduction of molecular components as reagents in blood tests for the identification of specific immunoglobulin (Ig)E radically changed the strategies used to identify the proper treatment of allergy. Along this line, Valenta et al.3 suggested selecting patients for AIT based on their reactivity to genuine molecules. More recently, Schmid-Grendelmeier4 showed that the best results obtained with AIT were achieved in patients sensitised to genuine components, while the worst were obtained in patients sensitised to cross-reacting components. Then, studies by Sastre et al.5,6 and Passalacqua et al.7 showed that AIT prescriptions were improved significantly by the availability of molecular allergy diagnosis (MAD) results. In particular, the list of allergens to be included in an AIT treatment was modified by the identification of sensitisation to a genuine allergen or of sensitisation to one or more cross-reacting components. As a result of these studies, Douladiris et al.8 described an algorithm that indicated the best AIT approach, which considered sensitisations to genuine and crossreacting components. Notably, this procedure also considered the possibility of false positives due to the presence of cross-reactive carbohydrate determinants (CCD) in the ‘natural highly purified’ component used in the assay. Along this line, a list of components to be re-evaluated following a direct test with CCD was given. Other important information was provided by Sastre et al.,9 who demonstrated that adverse reactions to AIT could be expected in patients characterised by a given IgE profile. Currently, in addition to molecular-based diagnoses, other novel approaches are being considered due to the support of artificial intelligence (AI), as well as the use of ‘big data’ analysis in the identification of specific IgE profiles in allergic patients. In this review, we describe these three approaches and consider both the pros and cons of an optimal and personalised diagnosis and AIT prescription.
THE USE OF MOLECULAR ALLERGY RESOURCES
Single-Plexed Diagnostics
Single-plexed diagnostic (SPD) tools have been implemented in a few different platforms (Phadia Immunosystem [Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA], Immulite [Siemens, Munich, Germany], and Euroline [Euroimmun UK Ltd., London, UK]). Even though their methods are slightly different, the results are comparable overall.10 The SPD approach is highly dependent on each physician’s habits and culture. Indeed, SPD is largely based on a physician’s specific requests; therefore, different strategies are possible. The simplest (which is also used in laboratory medicine) is the so-called ‘reflex test’, which is based on a list of recombinant molecules to be tested when a certain extractive agent is positive. According to this strategy, different choices can be made (Table 1) based on the depth of the investigation, costs, and reagent availability. Certainly, in the hands of a skilled allergist, the largest resource is the component selection and the deepest is the information. A different strategy is based on the doctor’s choice of components to be tested. This is based on an allergist’s experience and the complexity of the patient’s situation. Finally, a third approach provides the combined use of extractive allergens and components in the same panel. This is a more sophisticated approach that is based on the fact that a sensitisation can be defined as ‘genuine’ when the IgE recognises a genuine sensitiser molecular component. In this context, for instance, the use of birch raw extract should be avoided, and the Bet v 1 component should be used. Similarly, Parietaria spp. extractive allergens can be substituted by Par j 2, which is a genuine sensitising allergen. SPD can be used after exhaustive skin prick tests (SPT) and specific IgE (sIgE) testing when the clinical picture remains unclear and when AIT has to be prescribed. More recently, documented preparations for AIT have described that, in addition to short-term clinical improvements, the treatment can also have long-lasting effects.11 It is therefore crucial, when prescribing a long-lasting and expensive treatment, that the relevant molecular targets are clearly identified. However, when using this approach, one can only obtain what is sought. This can be seen as a real advantage or an embarrassing disadvantage.12
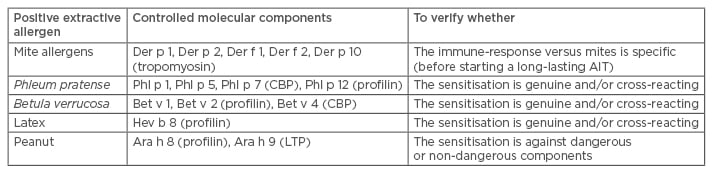
Table 1: Examples of reflex tests implemented in clinical pathology laboratories.
CBP: calcium-binding protein; LTP: lipid transfer protein; AIT: allergen immunotherapy.
Multiplexed Diagnostics
Multiplexed diagnostics (MPD) in allergy treatment are based on allergens (both highly purified molecules or recombinant components) inserted in a pre-defined diagnostic panel. For this reason, the list of molecules is based on different reasons, ranging from the real value in the diagnostic process, to the patent coverage, to the capacity of that given molecule to be linked to a solid phase.
The first platform available was ImmunoCAP ISAC (Thermo Fisher Scientific Inc.), which is now characterised by 112 different allergens (either purified or recombinant). The majority of the experience acquired in the multiplex diagnostic assay field came from the use of this tool.7,13
More recently, the MEDALL group, in strict co-operation with industries, developed a novel microarray by adding more than 70 new components to the standard ImmunoCAP ImmunoSolid Phase Allergy Chip (ISAC) 112 panel.14 The clinical features of the MEDALL microarray were further evaluated during the so-called ‘allergen-march’ from childhood to adolescence.15
A different approach has been developed by an English/Swedish company that designed a microarray involving both single-allergen components and whole extracts.16 This Microtest system was tested with three other allergy test methods (SPT, ImmunoCAP, and ImmunoCAP ISAC 112), and the results produced agreed with the currently used diagnostic tests.
Finally, another platform, which uses natural extracts and molecular components (ALEX® [Macro Array Diagnostics GmbH, Vienna, Austria]), seems to be a promising technological approach.
Allergen MPD offer both advantages and disadvantages. The advantages are represented by the large number of both natural purified and recombinant molecules on the same platform. For this reason, a small amount of blood is used, and the incubation and washing procedures are simple and rapid. More interestingly, sIgE MPD can also identify not only genuine and cross-reacting components but also harmless and potentially dangerous pan-allergens. Finally, the presence of more than one component of the same family (for example, profilins in ISAC are represented by four reagents, while in the Euroimmun, they are represented by two reagents) allows for clear specificity of the IgE repertoire. In this context, it is evident that if SPD obtains just what is sought, then MPD, which is based on a predefined allergen list, also gives unrequested results.10 This possibility has been considered a disadvantage by allergists; indeed, the occurrence of unexpected results may, in certain cases, embarrass the allergist. Other disadvantages are represented by the cost (SPD, which measures up to 10 molecules, is cheaper) and complexity of interpretation (see below).
Patients Obtaining Beneficial Effects From Molecular Allergy Diagnosis
Poly-sensitised patients can obtain beneficial effects from MAD for many reasons. Indeed, a patient is defined as poly-sensitised when more than two allergens belonging to different families are positive in SPT or sIgE testing. Therefore, a patient positive for mites, birch, and grasses is considered poly-sensitised. However, sensitisation to pan-allergens or cross-reacting components may influence the SPT and sIgE test results. For example, a patient who is sensitised to both Phl p 1 (a genuine grass component) and Phl p 12 (a profilin) may have positive results to many other allergens belonging to trees, grasses, and weeds. MAD, when used in such a patient, may not only indicate positivity to genuine components but also indicate that genuine components are negative and that profilin sensitisation is the cause of other positive results.
Food pollen syndromes are listed in Table 2. Pollen-food syndromes (as well as food-food and other complex food allergies) are caused by sensitisation to an inhalant component that cross-reacts with a very similar food component. The classic example is birch-apple syndrome, where sensitisation to Bet v 1 (a PR-10, the main component of birch sensitisation) is highly homologous to another PR-10 contained in apples, Mal d 1. Therefore, patients that eat apples have an oral allergy syndrome.17 Notably, for PR-10 sensitivity, heated, cooked, or industrially manipulated apples do not cause oral allergy syndrome. MAD allows for the identification of not only the principal sensitiser (the Bet v 1), but also for Mal d 1 sensitisation, which explains the observed syndrome. In contrast, other methods (such as SPT or sIgE with allergen extracts) do not allow for such a diagnosis. In this context, MAD is also useful in identifying foods that should be avoided if the allergist is planning to suggest a special diet for the patient.
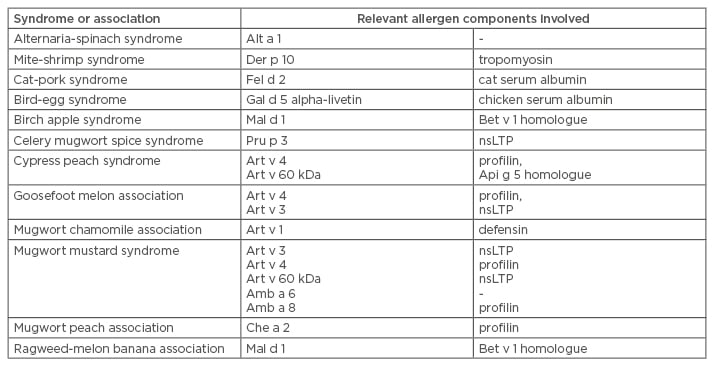
Table 2: Food pollen syndromes or associations.17
nsLTP: non-specific lipid transfer protein.
In a series of pivotal studies on grass sensitisation, Tripodi et al.18 showed that in a small percentage of cases, the IgE profile to Phleum components were represented in grass extracts for AIT. In particular, very few patients obtain an optimal vaccine according to their IgE repertoire. Additionally, the effects from the AIT can be foreseen on the basis of the IgE profile;19 however, an accurate IgE profile may suggest specific strategies in the AIT prescription.20 Another interesting and innovative use of allergen microarrays is allergen immunotherapy monitoring. Indeed, it was recently observed21 that allergen microarrays are useful in monitoring the development of allergen-specific IgG responses during specific immunotherapy, both against the allergen present in the specific immunotherapy vaccine as well as against cross- reactive allergens. The application of this technique may finally offer a general-purpose tool for monitoring the immunological effects of AIT, resulting in better treatment control and an even better understanding of therapeutically positive and negative results. These data were further supported by an article by Schmid et al.22 that demonstrated that pretreatment allergen component-specific IgE appears to determine IgG4 induction in the updosing phase. Induced IgG4 seems to suppress IgE levels in an ISAC, resulting in a marked decrease in ISAC-measured specific IgE levels after subcutaneous immunotherapy updosing. The authors concluded that decreased ISAC IgE levels can be used to monitor the blocking effect of allergen-specific immunotherapy-induced non-IgE antibodies.
MPD assays have been suggested as a helpful tool to predict the onset of adverse reactions, as shown by Sastre et al.,9 who documented that the adverse reaction rate, either local or systemic, is related to the number of sensitising grass pollen allergens.
COMPUTER ASSISTED DIAGNOSIS OF ALLERGY SENSITISATION
The introduction of expert system technology to support MAD introduced new diagnostic approach concepts. Indeed, the possible combinations of 112 allergens together with the difficulty of determining different component characteristics (and the reciprocal relationships) indicated that an AI tool could offer some advantages. In practice, relevant information can be obtained from AI elaborated allergen microarray results. Allergenius®, an expert system developed for ISAC result interpretation, by a team co-ordinated by Dr Melioli, can be considered a prototype because, by mixing different approaches, it offers a comprehensive view of the microarray results.23 ISAC seems to be redundant to some extent. For example, the number of profilins and lipid transfer proteins could be considered excessive. However, it was observed that a hierarchy of cross-reacting components can be identified using large-scale MAD assays.24 They showed that IgE reactivity to PR-10 proteins is characterised by a hierarchical intrarelationship: Bet v 1 > Mal d 1 > Cor a 1.04 > Ara h 8 > Pru p 1 > Aln g 1 > Api g 1 > Act d 8 > Gly m 4. For this reason, it is evident that many cross-reacting components are much more indicative than few. Along this line, Allergenius uses the rule that if the number of positive components is >40% of the total number of components of a given family, then the patient can be considered sensitised to the whole family of cross-reacting molecules. Additionally, Allergenius always identifies the first sensitiser of the cross-reacting component family as the member identified by the highest IgE score. Other added values can be derived from these rules. For example, if a discrepancy is identified between the SPT (or sIgE) results for a certain extractive allergen and the ISAC results (namely, a positive SPT result with negative specific components derived from that allergen), then Allergenius evaluates whether other crossreacting components (belonging to other allergen sources but cross-reacting with components well known to be detectable in the whole allergen) are also positive. Therefore, for example, if Ambrosia artemisiifolia is positive in the SPT but Amb a 1 is negative, other cross-reacting components are evaluated, such as profilins, PR-10, and CBP, which are all well represented in Ambrosia but not present in ISAC. If at least one of these cross-reacting components is positive, then there is no discrepancy. In contrast, if all the possible cross-reacting components are negative, then a clear discrepancy is declared. By using this approach, the number of apparent discrepancies is reduced significantly. Another added value described in Allergenius is related to the capacity of the expert system to evaluate the sums of the genuine inhalant and inhalant component scores that belong to cross-reacting families. As mentioned in the introduction, patients with genuine allergies seem to be more responsive to AIT, and Allergenius helps in the identification of these patients. Additionally, other issues can be managed by AI, including the identification of potentially dangerous sensitisations, the interpretation of sensitisations in certain geographic areas (such as lipid transfer proteins or peanut allergens in southern Europe), and the evaluation of the role of CCD in the interpretation of certain positivities (such as Phl p 4 and Jug r 2).
THE MANAGEMENT OF BIG DATASETS IN MOLECULAR ALLERGY
Big data are represented by statistical evaluation of a very large number of events. For example, traffic on the streets is measured in real time by aggregating data from cellular phones in that area. Certain tendencies on social behaviour are extracted from the very large number of data present in social networks. Therefore, extraction of relevant information from these large databases seems to be a frontier of laboratory epidemiology in the future. For example, the comparison of sensitisations with different components allows for better identifying areas where certain prophylactic activities (for example, tree cultivations or implants) may impact the quality of the life of patients. At present, big data are used to evaluate conditions in the supply chain of pharmacies for drugs and other medical devices. Alternatively, big data that are available on the internet allow for the identification of regions where certain pollens are present or where certain wind conditions may provide environments that are potentially harmful for an allergic patient. Data from microarrays, as well as data from large scale diagnostics laboratories, may occupy terabytes of memory. Furthermore, an accurate analysis of the sensitisation profile of large populations allows for molecular diagnostic management in a more convenient manner. From a clinical point of view, by using cluster analysis techniques to analyse data from thousands of ISAC sets,25 the presence of five different clusters of patients has been clearly shown. However, only Groups I and II (characterised by sensitisation to a large fraction of genuine components) are optimal targets for AIT, while Groups III and IV have worse expectations for success. Group V is constituted by food allergies. Such an analysis would have been impossible without techniques, such as cluster analysis, that allow for big data processing. Other approaches have been used in recent years. For example, in a specific environment represented by hymenoptera hypersensitisation of horses, Marti et al.26 described the use of advanced statistical methods to identify relevant sensitisations and validate their experimental approach. These techniques, which are particularly efficient when the variables are more frequent than the samples, allow for description of microarray features in a trustable manner. Prosperi et al.27 followed up on this approach and used machine learning to interpret allergen microarray results in relation to clinical symptoms. The results of these machine learning experiments will not only be extremely useful to allergists, but also to laboratory scientists who utilise single and multi-plexed diagnostics. Indeed, after validation, the mathematics used demonstrated reasonable discrimination between asthma, rhino-conjunctivitis, and wheezing, but not eczema (where it is well known that ISAC is largely negative). Therefore, the identification of certain patterns of specific IgE positivity allows for automatic assay result validation just by knowing some clinical aspects of the patient.
CONCLUSIONS
Allergists will face new challenges in the near future; these will include the availability of new analytical technologies that offer many more results to accurately describe the IgE repertoire of allergic patients. These new technologies, such as microarrays, produce a very large number of results that cannot be easily managed by allergists and are difficult to interpret for general practitioners. In these cases, AI can help in identifying the most relevant sensitisations, cross-reactions, and allergen lists that may have advantages for AIT. In the same context, many microarrays, which contain hundreds of results, will offer the possibility of studying sensitisation epidemiology with great accuracy. This will have an important impact not only on the management of single patients but also in large scale prophylaxis and allergy treatment strategies.






